Bacterial movement is an essential process influencing their ability to colonize new environments and interact with other organisms. In a study published in Science Advances, scientists from the LISM team “NMR of Molecular Assemblies,” in collaboration with Tâm Mignot’s team from LCB, have discovered the molecular mechanism by which the predatory bacterium, Myxococcus xanthus, regulates the assembly of proteins that enable its movement on surfaces as individual cells.
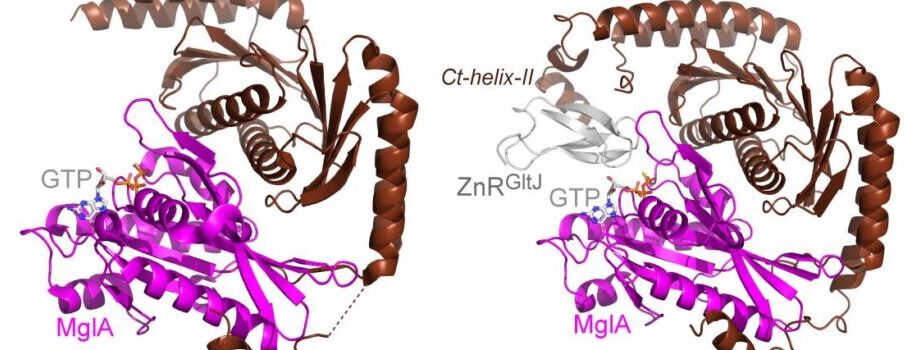
Cell motility universally relies on spatial regulation of focal adhesion complexes (FAs) connecting the substrate to cellular motors. In bacterial FAs, the Adventurous gliding motility machinery (Agl-Glt) assembles at the leading cell pole following a Mutual gliding-motility protein (MglA)-guanosine 5′-triphosphate (GTP) gradient along the cell axis. Here, we show that GltJ, a machinery membrane protein, contains cytosolic motifs binding MglA-GTP and AglZ and recruiting the MreB cytoskeleton to initiate movement toward the lagging cell pole. In addition, MglA-GTP binding triggers a conformational shift in an adjacent GltJ zinc-finger domain, facilitating MglB recruitment near the lagging pole. This prompts GTP hydrolysis by MglA, leading to complex disassembly. The GltJ switch thus serves as a sensor for the MglA-GTP gradient, controlling FA activity spatially.
Attia B, My L, Castaing JP, Dinet C, Le Guenno H, Schmidt V, Espinosa L, Anantharaman V, Aravind L, Sebban-Kreuzer C, Nouailler M, Bornet O, Viollier P, *Elantak L, *Mignot T.
Science Advances, 2024 May 31, DOI : 10.1126/sciadv.adn2789
Discover the article here : http://A molecular switch controls assembly of bacterial focal adhesions

This article is also featured in a scientific news update from the CNRS Institute of Biology:
Published on 22/07/2024
