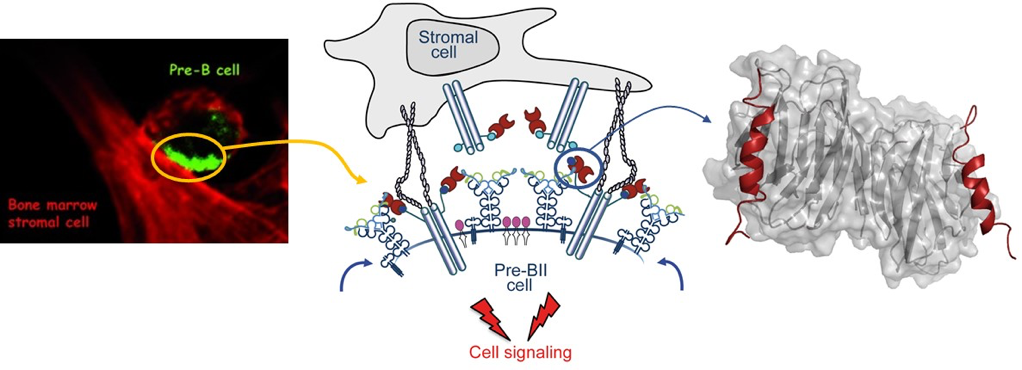
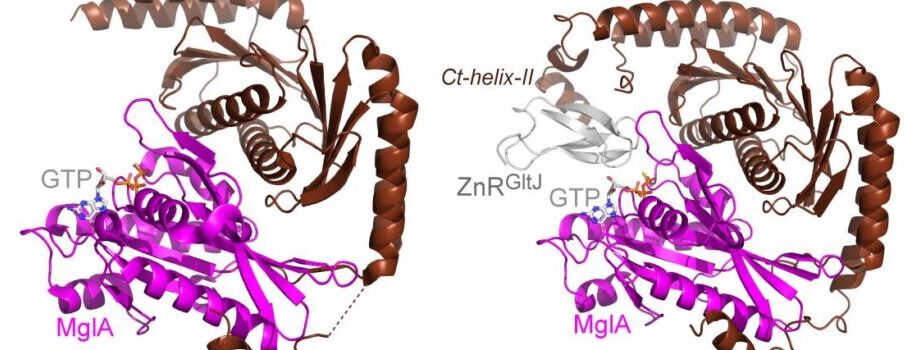
Understanding the structural basis of protein assemblies that are functionally important for various aspects of cell contacts with its microenvironment.
Cell contacts with the microenvironment orchestrate the development of organisms, homeostasis, and the functions of individual cells. When cells do not interact correctly or decode external molecular messages incorrectly, cellular responses are disturbed, and diseases can develop.
Therefore, understanding the structural basis of protein assemblies involved in the sophisticated mechanisms of action developed by cells to interact/communicate with their microenvironment is of main importance.
Our research is focused on the study of these membrane-associated protein complexes. The primary focus of our research is on how human Galectins interactions are regulated at the cell surface during cell-cell interactions.
This study involves protein-protein as well as protein-carbohydrates interactions. We also seek to understand the mechanisms regulating bacterial cell motility from structural studies of focal adhesion protein complexes in M. xanthus.
To pursue our goals, we use Nuclear Magnetic resonance (NMR) spectroscopy combined with biochemical and biophysical techniques in an integrated structural biology approach. NMR can provide an atomic-level view of the structure and dynamics of macromolecular systems in solution. A particular strength of NMR is its ability to reveal the structural details of specific interactions between biological macromolecules, even in the case of transient complexes between proteins and their targets in solution.
Overall, our aim is to decipher at atomic level the assembly of multi-protein complexes, determine their structures, gain insight into their activities and regulation, and identify roles for proteins or domains of unknown function.
Members
Researchers
- Françoise Guerlesquin (Past team leader)
Post doctoral researchers
- Bouchra Attia
PhD students
- Bouchra Attia
- Pauline Touarin
- Romain Navarro
Technical staff
- Audrey Courbois
Master 2 students
- Chloe Klincewicz
Undergraduates
- Hanane Laalioui (M1)
- Clara Hoffmann (Poly-Tech)
Publications
A molecular switch controls assembly of bacterial focal adhesions
Bouchra Attia, Laetitia My, Jean Philippe Castaing, Céline Dinet, Hugo Le Guenno, Victoria Schmidt, Leon Espinosa, Vivek Anantharaman, L. Aravind, Corinne Sebban-Kreuzer, Matthieu Nouailler, Olivier Bornet, Patrick Viollier, Latifa Elantak, Tâm Mignot
Science Advances 10 (2024)10.1126/sciadv.adn2789
Pre-B cell receptor acts as a selectivity switch for galectin-1 at the pre-B cell surface
Pauline Touarin, Bastien Serrano, Audrey Courbois, Olivier Bornet, Qian Chen, Lincoln G Scott, James R Williamson, Corinne Sebban-Kreuzer, Stéphane J.C. Mancini, Latifa Elantak
Cell Reports 43:114541 (2024)10.1016/j.celrep.2024.114541
1H, 13C and 15N chemical shift assignments of the ZnR and GYF cytoplasmic domains of the GltJ protein from Myxococcus xanthus
Bouchra Attia, Bastien Serrano, Olivier Bornet, Françoise Guerlesquin, Laetitia My, Jean-Philippe Castaing, Tâm Mignot, Latifa Elantak
Biomolecular NMR Assignments 16:219-223 (2022)10.1007/s12104-022-10083-6
FrzS acts as a polar beacon to recruit SgmX , a central activator of type IV pili during Myxococcus xanthus motility
Sarah Bautista, Victoria Schmidt, Annick Guiseppi, Emillia M F Mauriello, Bouchra Attia, Latifa Elantak, Tâm Mignot, Romain Mercier
EMBO Journal e111661 (2022)10.15252/embj.2022111661
Identification of the three zinc-binding sites on tau protein
Romain Rocca, Philipp O Tsvetkov, Andrey Golovin, Diane Allegro, Pascale Barbier, Soazig Malesinski, Françoise Guerlesquin, François Devred
International Journal of Biological Macromolecules 209:779-784 (2022)10.1016/j.ijbiomac.2022.04.058
Glutamate optimizes enzymatic activity under high hydrostatic pressure in Desulfovibrio species: effects on the ubiquitous thioredoxin system
Hélène Gaussier, M. Nouailler, Edouard Champaud, Edwige Garcin, Sebban -Kreuzer, Olivier Bornet, Marc Garel, Christian Tamburini, Laetitia Pieulle, Alain Dolla, Nathalie Pradel
Extremophiles 25:385-392 (2021)10.1007/s00792-021-01236-x
Chemokines modulate glycan binding and the immunoregulatory activity of galectins
Lucía Sanjurjo, Iris Schulkens, Pauline Touarin, Roy Heusschen, Ed Aanhane, Kitty Castricum, Tanja de Gruijl, Ulf Nilsson, Hakon Leffler, Arjan Griffioen, Latifa Elantak, Rory Koenen, Victor Thijssen
Communications Biology 4:1415 (2021)10.1038/s42003-021-02922-4
The unusual structure of Ruminococcin C1 antimicrobial peptide confers clinical properties
Clarisse Roblin, Steve Chiumento, Olivier Bornet, Matthieu Nouailler, Christina S. Müller, Katy Jeannot, Christian Basset, Sylvie Kieffer-Jaquinod, Yohann Couté, Stéphane Torelli, Laurent Le pape, Volker Schünemann, Hamza Olleik, Bruno de La Villeon, Philippe Sockeel, Eric Di Pasquale, Cendrine Nicoletti, Nicolas Vidal, Leonora Poljak, Olga Iranzo, Thierry Giardina, Michel Fons, Estelle Devillard, Patrice Polard, Marc Maresca, Josette Perrier, Mohamed Atta, Francoise Guerlesquin, Mickaël Lafond, Victor Duarte
Proceedings of the National Academy of Sciences of the United States of America 117:19168-19177 (2020)10.1073/pnas.2302405120
Deciphering the specific interaction between the acyl carrier protein IacP and the T3SS‐major hydrophobic translocator SipB from Salmonella
Mickaël J Canestrari, Bastien Serrano, Julia Bartoli, Valerie Prima, Olivier Bornet, Rémy Puppo, Emmanuelle Bouveret, Francoise Guerlesquin, Julie Viala
FEBS Letters (2019)10.1002/1873-3468.13593
Ruminococcin C, a promising antibiotic produced by a human gut symbiont
Steve Chiumento, Clarisse Roblin, Sylvie Kieffer-Jaquinod, Sybille Tachon, Chloé Leprêtre, Christian Basset, Dwi Aditiyarini, Hamza Olleik, Cendrine Nicoletti, Olivier Bornet, Olga Iranzo, Marc Maresca, Renaud Hardré, Michel Fons, Thierry Giardina, Estelle Devillard, Françoise Guerlesquin, Yohann Couté, Mohamed Atta, Josette Perrier, Mickael Lafond, Victor Duarte
Science Advances 5:eaaw9969 (2019)10.1126/sciadv.aaw9969
Electrostatic interactions between the CTX phage minor coat protein and the bacterial host receptor TolA drive the pathogenic conversion of Vibrio cholerae
Laetitia Houot, Romain Navarro, Matthieu Nouailler, Denis Duché, Francoise Guerlesquin, Roland Lloubès
Journal of Biological Chemistry 292:13584-13598 (2017)10.1074/jbc.M117.786061
1H, 13C and 15N assignments of the C-terminal intrinsically disordered cytosolic fragment of the receptor tyrosine kinase ErbB2
Yinghui Wang, Louise Pinet, Nadine Assrir, Latifa Elantak, Francoise Guerlesquin, Ali Badache, Ewen Lescop, Carine van Heijenoort
Biomolecular NMR Assignments 12:23-26 (2017)10.1007/s12104-017-9773-4
(1)H, (15)N and (13)C resonance assignments of the C-terminal domain of Vibrio cholerae TolA protein.
Romain Navarro, Olivier Bornet, Laetitia Houot, Roland Lloubes, Francoise Guerlesquin, Matthieu Nouailler
Biomol NMR Assign (2016)10.1007/s12104-016-9690-y
Pre-B cell receptor binding to galectin-1 modifies galectin-1/carbohydrate affinity to modulate specific galectin-1/glycan lattice interactions
Jeremy Bonzi, Olivier Bornet, Stéphane Betzi, Brian T Kasper, Lara K Mahal, Stéphane Mancini, Claudine Schiff, Corinne Sebban-Kreuzer, Francoise Guerlesquin, Latifa Elantak
Nature Communications 6:6194 (2015)10.1038/ncomms7194
Examination of galectin-induced lattice formation on early B-cell development.
Stephane J C Mancini, Latifa El Antak, Annie Boned, Marion Espeli, Francoise Guerlesquin, Claudine Schiff
Methods in Molecular Biology 1207:169-84 (2015)10.1007/978-1-4939-1396-1_11
Identification of a Src kinase SH3 binding site in the C-terminal domain of the human ErbB2 receptor tyrosine kinase.
Olivier Bornet, Matthieu Nouailler, Michaël Feracci, Corinne Sebban-Kreuzer, Deborah Byrne, Hubert Halimi, Xavier Morelli, Ali Badache, Francoise Guerlesquin
FEBS Letters 588:2031-6 (2014)10.1016/j.febslet.2014.04.029
1H, 13C and 15N backbone and side-chain chemical shift assignments for reduced unusual thioredoxin Patrx2 of Pseudomonas aeruginosa.
Edwige B Garcin, Olivier Bornet, Latifa El Antak, Matthieu Nouailler, Francoise Guerlesquin, Corinne Sebban-Kreuzer
Biomol NMR Assign 8:247-50 (2014)10.1007/s12104-013-9493-3
Glycan Dependence of Galectin-3 Self-Association Properties
Hubert Halimi, Annafrancesca Rigato, Deborah Byrne, Géraldine Ferracci, Corinne Sebban-Kreuzer, Latifa Elantak, Francoise Guerlesquin
PLoS ONE 9:e111836 (2014)10.1371/journal.pone.0111836
Structural Basis for Galectin-1-dependent Pre-B Cell Receptor (Pre-BCR) Activation
Latifa Elantak, Marion Espeli, Annie Boned, Olivier Bornet, Jeremy Bonzi, Laurent Gauthier, Mikael Feracci, Philippe Roche, Francoise Guerlesquin, Claudine Schiff
Journal of Biological Chemistry 287:44703-44713 (2012)10.1074/jbc.m112.395152
DnaJ (Hsp40 Protein) Binding to Folded Substrate Impacts KplE1 Prophage Excision Efficiency
Tania Puvirajesinghe, Latifa Elantak, Sabrina Lignon, Nathalie Franche, Marianne Ilbert, Mireille Ansaldi
Journal of Biological Chemistry 287:14169-14177 (2012)10.1074/jbc.M111.331462
The Indispensable N-Terminal Half of eIF3j/HCR1 Cooperates with its Structurally Conserved Binding Partner eIF3b/PRT1-RRM and with eIF1A in Stringent AUG Selection
Latifa Elantak, Susan Wagner, Anna Herrmannová, Martina Karásková, Edit Rutkai, Peter J Lukavsky, Leoš Valášek
Journal of Molecular Biology 396:1097-1116 (2010)10.1016/j.jmb.2009.12.047
Structure of eIF3b RNA Recognition Motif and Its Interaction with eIF3j
Latifa Elantak, Andreas G Tzakos, Nicolas Locker, Peter J Lukavsky
Journal of Biological Chemistry 282:8165-8174 (2007)10.1074/jbc.m610860200
Collaborations
- National
- Tâm Mignot, LCB UMR7283, Marseille
- Romain Mercier, LCB UMR7283, Marseille
- Stephane Mancini, Rennes
- Guido Pintacuda, Lyon
- International
- Victor Thijssen, Amsterdam
- Daniel Abankwa, Luxembourg
- Lincoln Scott, San Diego
Funding