The “Sensing environment & community lifestyle” team is studying the regulatory pathways
that control biofilm formation in P. aeruginosa as well as the processes of evolution and
adaptation that this bacterium undergoes in this lifestyle.
90% of the microbiological activity in an ecosystem (aquatic environment, soil, human body, etc.) comes for the most part from sedentary flora adhering to surfaces in a structure called biofilm, which is an aggregate of micro-organisms (mostly bacteria), fixed to a surface, and stuck in a self-produced gangue of exopolymers.
The presence of biofilms is increasingly identified as the recurring source of both public health and industrial problems, which are often serious and sometimes dramatic. Pseudomonas aeruginosa is a gram-negative bacterium, an opportunistic human pathogen with a very high adaptive capacity since it can colonise many ecological niches.
Practically harmless in healthy people, this Gram-negative environmental bacillus can become a deadly threat to immunodeficient individuals, intubated-ventilated patients in intensive care units or those suffering from chronic diseases such as cystic fibrosis.
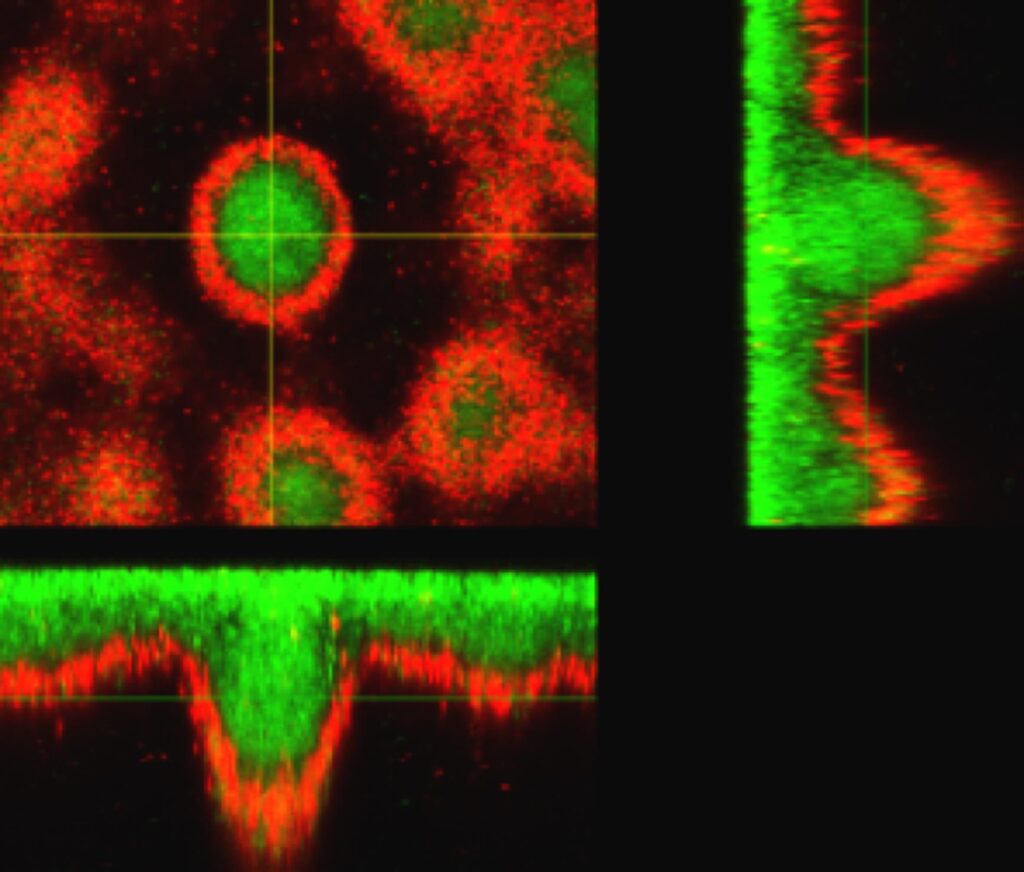
During chromic infections, P. aeruginosa adopts a biofilm lifestyle that allows it to evade the immune system, limits the penetration of any antibiotic treatment and promotes the emergence of specific resistances.
The Sensing environment & community lifestyle team studies, using genomic, fluorescent microscopy and structural biology the regulatory pathways that trigger the formation of the biofilm and the evolutionary and adaptive processes that P. aeruginosa undergoes in its lifestyle. Our aim is to propose alternative therapeutic strategies for the treatment of P. aeruginosa infections.
Members
- Gaël Chambonnier : PhD student
- Firas Fadel : Post-Doc
- Coralie Marchand : Master 2 MBVB
- Gianlucca Nicastro : Post-Doc
- David Redelberger : Technical staff
- Sarah Bigot : Researcher
- Nicolas Roux : PhD student
- Sophie de Bentzmann : INSERM Researcher
- Lorène Roux : PhD student
- Jennifer Spagnolo : PhD student
- David Redelberger : Technical staff
- Christophe Bernard : PhD student
- Caroline Giraud : PhD student
- Friederike Ewald : Master 2 BBSG
- Laetitia Houot : Post Doc
- Segolène Ruer : PhD student
Publications
Pre-B cell receptor acts as a selectivity switch for galectin-1 at the pre-B cell surface
Pauline Touarin, Bastien Serrano, Audrey Courbois, Olivier Bornet, Qian Chen, Lincoln G Scott, James R Williamson, Corinne Sebban-Kreuzer, Stéphane J.C. Mancini, Latifa Elantak
Cell Reports 43:114541 (2024)10.1016/j.celrep.2024.114541
Unravelling the molecular basis of metabolic interactions in synthetic consortia dedicated for bio-H2 production
Louis Delecourt, Magali Roger, Pascale Infossi, Christophe Bordi, Marie-Thérèse Giudici-Orticoni
Biochimica biophysica acta (BBA) - Bioenergetics 1863:148825 (2022)10.1016/j.bbabio.2022.148825
The horizontal transfer of Pseudomonas aeruginosa PA14 ICE PAPI-1 is controlled by a transcriptional triad between TprA, NdpA2 and MvaT
Gauthier Dangla Pélissier, Nicolas Roux, Victoria Schmidt, Gaël Chambonnier, Moly Ba, Corinne Sebban-Kreuzer, Sophie de Bentzmann, Caroline Giraud, Christophe Bordi
Nucleic Acids Research 49:10956-10974 (2021)10.1093/nar/gkab827
Editorial: Gram-Negative Pathogenesis
Hidetada Hirakawa, Christophe Bordi, Haruyoshi Tomita
Frontiers in Microbiology 12 (2021)10.3389/fmicb.2021.813062
Connected partner-switches control the life style of Pseudomonas aeruginosa through RpoS regulation
Sophie Bouillet, Moly Ba, Laetitia Houot, Chantal Iobbi-Nivol, Christophe Bordi
Scientific Reports 9 (2019)10.1038/s41598-019-42653-5
Constitutive activation of MexT by amino acid substitutions results in MexEF-OprN overproduction in clinical isolates of Pseudomonas aeruginosa
Paulo Juarez, Isabelle Broutin, Christophe Bordi, Patrick Plesiat, Catherine Llanes
Antimicrobial Agents and Chemotherapy (2018)10.1128/AAC.02445-17
NMR assignments of the GacS histidine-kinase periplasmic detection domain from Pseudomonas aeruginosa PAO1
Ahmad Ali-Ahmad, Olivier Bornet, Firas Fadel, Yves Bourne, Florence Vincent, Christophe Bordi, Françoise Guerlesquin, Corinne Sebban-Kreuzer
Biomolecular NMR Assignments 11:25-28 (2017)10.1007/s12104-016-9714-7
Structural and functional insights into the periplasmic detector domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa
Ahmad Ali-Ahmad, Firas Fadel, Corinne Sebban-Kreuzer, Moly Ba, Gauthier Dangla Pélissier, Olivier Bornet, Françoise Guerlesquin, Yves Bourne, Christophe Bordi, Florence Vincent
Scientific Reports 7:11262 (2017)10.1038/s41598-017-11361-3
Cyclic-di-GMP regulates lipopolysaccharide modification and contributes to Pseudomonas aeruginosa immune evasion Europe PMC Funders Group
Ronan R Mccarthy, Maria J Mazon-Moya, Joana A Moscoso, Youai Hao, Joseph S Lam, Christophe Bordi, Serge Mostowy, Alain Filloux
Nature Microbiology 2:17027 (2017)10.1038/nmicrobiol.2017.27
The Hybrid Histidine Kinase LadS Forms a Multicomponent Signal Transduction System with the GacS/GacA Two-Component System in Pseudomonas aeruginosa
Gaël Chambonnier, Lorène Roux, David Redelberger, Firas Fadel, Alain Filloux, Melissa Sivaneson, Sophie de Bentzmann, Christophe Bordi
PLoS Genetics 12:e1006032 (2016)10.1371/journal.pgen.1006032
M-CSF improves protection against bacterial and fungal infections after hematopoietic stem/progenitor cell transplantation
Prashanth Kandalla, Sandrine Sarrazin, Kaaweh Molawi, Carole Berruyer, David Redelberger, Anne Favel, Christophe Bordi, Sophie de Bentzmann, Michael H. Sieweke
Journal of Experimental Medicine 213:2269-2279 (2016)10.1084/jem.20151975
Pre-B cell receptor binding to galectin-1 modifies galectin-1/carbohydrate affinity to modulate specific galectin-1/glycan lattice interactions
Jeremy Bonzi, Olivier Bornet, Stéphane Betzi, Brian T Kasper, Lara K Mahal, Stéphane Mancini, Claudine Schiff, Corinne Sebban-Kreuzer, Francoise Guerlesquin, Latifa Elantak
Nature Communications 6:6194 (2015)10.1038/ncomms7194
Construction of Pseudomonas aeruginosa two-hybrid libraries for high-throughput assays.
Sophie De Bentzmann, Christophe Bordi
Methods in Molecular Biology 1149:225-41 (2014)10.1007/978-1-4939-0473-0_19
Construction of a Pseudomonas aeruginosa genomic DNA library.
Christophe Bordi
Methods in Molecular Biology 1149:555-63 (2014)10.1007/978-1-4939-0473-0_42
Identification of a Src kinase SH3 binding site in the C-terminal domain of the human ErbB2 receptor tyrosine kinase.
Olivier Bornet, Matthieu Nouailler, Michaël Feracci, Corinne Sebban-Kreuzer, Deborah Byrne, Hubert Halimi, Xavier Morelli, Ali Badache, Francoise Guerlesquin
FEBS Letters 588:2031-6 (2014)10.1016/j.febslet.2014.04.029
1H, 13C and 15N backbone and side-chain chemical shift assignments for reduced unusual thioredoxin Patrx2 of Pseudomonas aeruginosa.
Edwige B Garcin, Olivier Bornet, Latifa El Antak, Matthieu Nouailler, Francoise Guerlesquin, Corinne Sebban-Kreuzer
Biomol NMR Assign 8:247-50 (2014)10.1007/s12104-013-9493-3
Glycan Dependence of Galectin-3 Self-Association Properties
Hubert Halimi, Annafrancesca Rigato, Deborah Byrne, Géraldine Ferracci, Corinne Sebban-Kreuzer, Latifa Elantak, Francoise Guerlesquin
PLoS ONE 9:e111836 (2014)10.1371/journal.pone.0111836
Pseudomonas aeruginosa Genome Evolution in Patients and under the Hospital Environment.
Céline Lucchetti-Miganeh, David Redelberger, Gaël Chambonnier, François Rechenmann, Sylvie Elsen, Christophe Bordi, Katy Jeannot, Ina Attrée, Patrick Plésiat, Sophie De Bentzmann
Pathogens 3:309-40 (2014)10.3390/pathogens3020309
Structural and Functional Characterization of the Clostridium perfringens N-Acetylmannosamine-6-phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
Marie-Cécile Pélissier, Corinne Sebban-Kreuzer, Françoise Guerlesquin, James A Brannigan, Yves Bourne, Florence Vincent
Journal of Biological Chemistry 289:35215-35224 (2014)10.1074/jbc.m114.604272
A gacS deletion in Pseudomonas aeruginosa cystic fibrosis isolate CHA shapes its virulence
Khady Mayebine Sall, Maria Guillermina Casabona Casabona, Christophe Bordi, Philippe Huber, Sophie De Bentzmann, Ina Attrée, Sylvie Elsen
PLoS ONE 9:e95936 (2014)10.1371/journal.pone.0095936
Unique biofilm signature, drug susceptibility and decreased virulence in Drosophila through the Pseudomonas aeruginosa two-component system PprAB.
Sophie de Bentzmann, Caroline Giraud, Christophe S. Bernard, Virginie Calderon, Friederike Ewald, Patrick Plésiat, Cathy Nguyen, Didier Grunwald, Ina Attree, Katy Jeannot, Marie-Odile Fauvarque, Christophe Bordi
PLoS Pathogens 8:e1003052 (2012)10.1371/journal.ppat.1003052
A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa.
Laetitia Houot, Adeline Fanni, Sophie De Bentzmann, Christophe Bordi
Microbiology (Reading, Engl.) 158:1964-71 (2012)10.1099/mic.0.057059-0
Development of a genetic tool for activating chromosomal expression of cryptic or tightly regulated loci in Pseudomonas aeruginosa.
Jennifer Spagnolo, Sarah Bigot, Yann Denis, Christophe Bordi, Sophie De Bentzmann
Plasmid 67:245-51 (2012)10.1016/j.plasmid.2011.12.006
Hacking into bacterial biofilms: a new therapeutic challenge.
Christophe Bordi, Sophie De Bentzmann
Annals of Intensive Care 1:19 (2011)10.1186/2110-5820-1-19
1H, 13C and 15N chemical shift assignments of the thioredoxin from the obligate anaerobe Desulfovibrio vulgaris Hildenborough.
Edwige B Garcin, Olivier Bornet, Laetitia Pieulle, Françoise Guerlesquin, Corinne Sebban-Kreuzer
Biomol NMR Assign 5:177-9 (2011)10.1007/s12104-011-9294-5
Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression.
Melissa Sivaneson, Helga Mikkelsen, Isabelle Ventre, Christophe Bordi, Alain Filloux
Mol. Microbiol. 79:1353-66 (2011)10.1111/j.1365-2958.2010.07527.x
Distinct oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand binding site
Florence Vincent, Adam Round, Aline Reynaud, Christophe Bordi, Alain Filloux, Yves Bourne
Environmental Microbiology 12:1775-1786 (2009)10.1111/j.1462-2920.2010.02264.x
Collaborations
- National
- Dr Chantal Iobbi-Nivol, BIP, Marseille
- Dr Marie Thérèse Giudici-Orticoni, BIP, Marseille
Funding
















