James Sturgis’ research group is interested in the dynamics and assembly of membrane proteins. We are interested in how membrane proteins fold and assemble in the membrane to form both stable structures, such as the photosynthetic apparatus, or transient dynamic structures.
To investigate the assembly of membrane proteins we use a combination of different theroetical and experimental techniques. In a series of numerical approaches we are using computer simulations and theoretical analysis to investigate the assembly process at various levels of detail (atomistic, coarse-grained and ultra-coarse grained). While at the experimental level we use spectroscopic, microscopic, molecular biology and synthetic biology methods to analyse the assembly process.
We have a series of biological systems that we investigate using these techniques: AqpZ, the Bacterial photosynthetic apparatus and mitochondrial anion channel, VDAC.
1 – AqpZ assembly
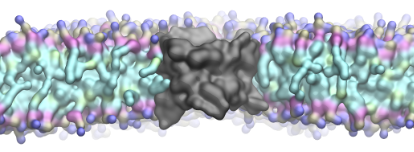
The membrane protein Aquaporin Z (AqpZ), from Escherichia coli forms a tetrameric water channel. However very little is known about the rules that govern membrane protein folding and assembly. We hope to be able to decode some of these rules by examining different steps in the folding of this protein.
Our research focusses on understanding the interactions between the protein and membrane lipids and in particular aspects of the protein surface and the lipid composition that modulate formation of tetrameric complexes and their subsequent assembly into 2D crystals. For this we are using various biochemical and biophysical techniques in the laboratory (fluorescence, FTIR spectroscopy, EM, lipidomics) or in collaboration with leading laboratories around the world (nmr, AFM and single molecule measurements).
in a lipid membrane (Coll. S. Scheuring).
2 – Organization of the bacterial photosynthetic apparatus
A larger system we are studying is the bacterial photosynthetic system. This simple system forms specialized membranes composed essentially of 3 to 4 intergal membrane complexes. The different proteins are able to assemble spontaneously into specialise membranes within the bacteria, the morphology and composition of these domains are important for function, and we try to understand why certain proteins assemble into these domains and how their internal structure is determined.
Currently we are using multiple methods to modify the organization of membrane components, using molecular biology tools to modulate expression of the different components in living bacteria and constructing artificial photosynthetic systems bottom up. In both cases we study the organization and function of the system in order to elucidate the links between structure and function and the robustness of this simple integrated biological system.
3 – Understanding the role of the Voltage-Dependent Anion Channel (VDAC) in mitochondrial regulation
VDAC channels are the most abundant proteins in the mitochondrial outer membrane (MOM) and consist of 19-stranded β-barrels that facilitate the passage of ions and metabolites, such as ATP and ADP, in and out of the mitochondria. In addition to their transport function, VDAC channels play a crucial role in various mitochondrial regulatory pathways, interacting with partner proteins involved in processes like apoptosis, neurodegeneration, and calcium homeostasis. Despite their critical importance in mitochondrial regulation, the specific mechanisms of how VDAC channels interact with these partners remain poorly understood.
Our team employs a multidisciplinary approach, combining biochemistry, biophysics techniques (e.g., EPR, AFM), and molecular dynamic simulations to deepen our understanding of VDAC’s role, particularly focusing on the following questions:
a) VDAC Isoform specificity
In mammals, there are three VDAC isoforms—VDAC1, VDAC2, and VDAC3—with approximately 80% sequence similarity and broad, overlapping tissue distribution. While all isoforms function in metabolite and ion transport, their distinct physiological roles remain largely unclear. A key focus of our research is understanding how each isoform selects its protein partners and interacts with lipids, a critical yet poorly understood aspect of their functional diversity and role in mitochondrial physiology.
b) VDAC assemblies and the role of lipids
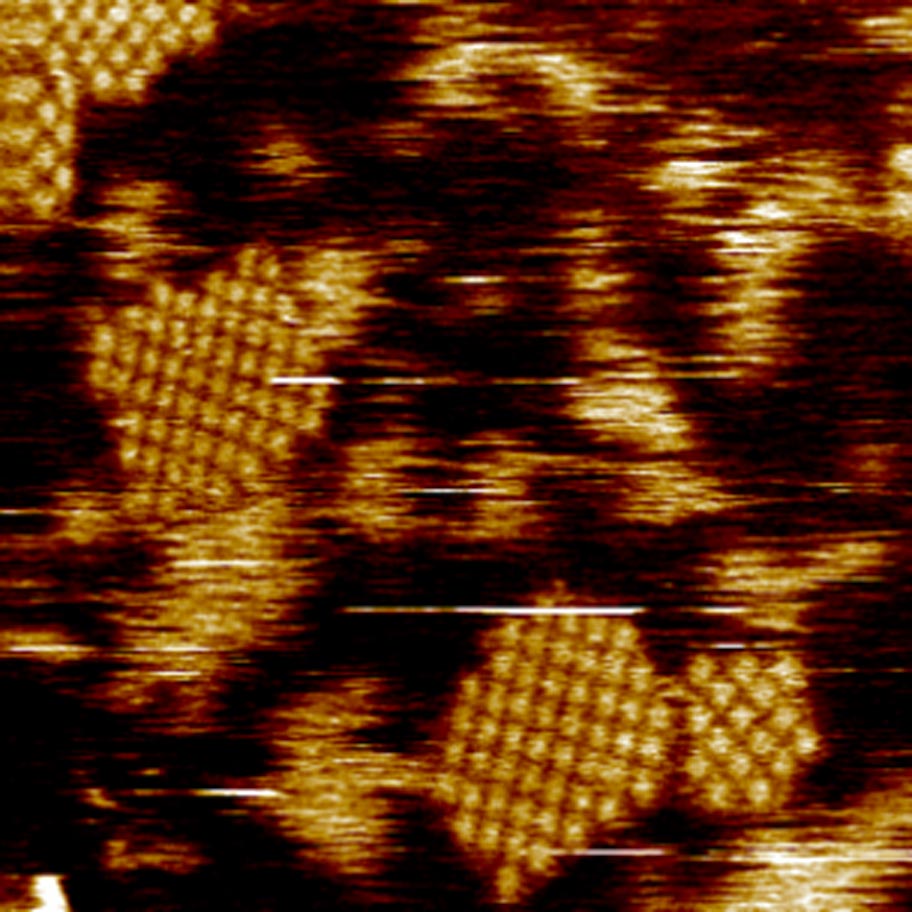
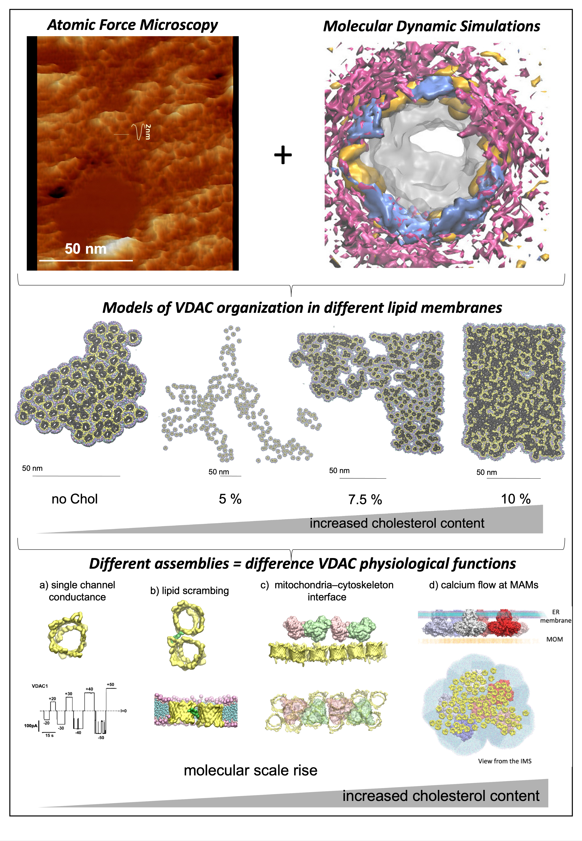
Mitochondrial physiology is intricately linked to the oligomerization of VDAC. However, the molecular determinants of VDAC oligomerization remain poorly understood. We investigate the effects of lipids of the Mitochondrial Outer Membrane (MOM) on VDAC assemblies, observing that VDAC forms lipid-sensitive clusters, which compaction is regulated by cholesterol. We are using a combination of biochemistry, AFM and molecular dynamics simulations and modeling to understand the molecular forces driving the different isoforms’ assemblies.
We employed an integrative approach, combining molecular dynamics simulations and AFM of VDAC in bilayers with varying lipid compositions, to demonstrate that lipids govern VDAC organization. This underscores the profound impact of the lipid environment on VDAC function, triggering different level of assembly and compaction of VDAC, which can be linked to different function of the protein. This highlights the physiological significance of lipid heterogeneity and changes within biological membranes—whether arising from membrane contacts or pathologies—in modulating VDAC behavior.
c) VDAC interaction with apoptotic proteins
Apoptosis, or “cell suicide,” is a vital process whose dysfunction can lead to various disorders, including cancer and Alzheimer’s disease. Mitochondria-mediated apoptosis involves the permeabilization of the mitochondrial outer membrane (MOM), a key event regulated by Bcl-2 family proteins. In this process, the pro-apoptotic proteins Bax and Bak are activated and integrate into the MOM to form pores. VDAC2 is essential for Bax- and Bak-induced MOM permeabilization, yet the exact mechanism of pore formation remains unclear. Notably, the lack of structural data on the VDAC2-Bax interaction limits our understanding of how this process is regulated, and how other proteins or small molecules might stabilize or disrupt the complex to influence apoptosis. Despite knowing the key players, the precise mechanism behind this critical event in cell fate remains an enigma.
Using a combination of biochemical and biophysical techniques, our team aims to reconstitute and functionnally a
Members
Past members
Jérôme Hénin (http://www-lbt.ibpc.fr/people/henin)
Pierre Hubert 2004-2014
Katia Duquesne 2009-2012
Cecile Blanchard 2010-2012
PhD Students
Batiste Thienpont 2018-2023
Sevde Karatas 2018-2022
Marcel Giansily 2017-2022
Marlon Sidore 2014-2018
Victoria Schmidt 2013-2017 (https://lism.imm.cnrs.fr/member/victoria-schmidt)
Wang Dong 2013-2017
Paul Sawma 2009-2013
Jonathan Khao 2007-2011 (https://clarafi.com/instructors/jonathan-khao)
Astrid Wahl 2006-2010
Camille Mascle-Allemand 2004-2008
Jaime Arce-Lopera 2003-2007
Undergraduates
2024 Manon Banegas
2024 Clara Segura
2024 Guillaume Hernitte
2023 Gary Mazoyer
2023 Geoffrey Dassonville
2023 Elissa Chams
2023 Ibrahima Faye
2023 Melanie Lucas Carrico
2023 Raime Zerdoum
2022 Angelica Lopez
2021 Adel Beghiah
2020 Wafa Benmenour
2020 Roumeissa Zouiher
2020 Nafisatou Drame
2019 Jennifer Arrindell
2019 Maxime Armand
2018 Axel Linden
2018 Camille Garcia
2016 Laurie Nigri
2016 Jamal Saad
Publications
Atypical thioredoxin Patrx2 enhances alginate production in mucoid Pseudomonas aeruginosa
Marie M Grandjean, James N Sturgis, Edwige B Garcin, Moly Ba, Olivier Bornet, Christophe Bordi, Latifa Elantak, Corinne Sebban-Kreuzer
Redox Biology 90:104010 (2026)10.1016/j.redox.2026.104010
Interfacial effects break the canonical permeability-selectivity trade-off in biological nanopores
María Queralt-Martín, Laidy M Alvero-Gonzalez, Marcel Aguilella-Arzo, D. Aurora Perini, Lucie A Bergdoll, Antonio Alcaraz
Journal of Colloid and Interface Science 703:139266 (2026)10.1016/j.jcis.2025.139266
Crystal Digital PCR™ Enables Precise Quantification of Species Abundance in Microbial Mixtures
Louis Delecourt, Mila Reset, Lionel Bertaux, James Sturgis, Yann Denis, Marie-Thérèse Giudici-Orticoni, Magali Roger, Christophe Bordi
Microorganisms 13:2592 (2025)10.3390/microorganisms13112592
Anionic Lipids Regulate the Light-Harvesting Complex 1-Reaction Center Photocycle in Purple Bacteria
Olivia Fiebig, Graham Schmidt, Neetu Singh Yadav, Dihao Wang, Muath Nairat, Helen Tang, Yi Ji, Valérie Prima, James N. Sturgis, Josh Vermaas, Dvir Harris, Gabriela Schlau-Cohen
Journal of the American Chemical Society (2025)10.1021/jacs.5c12301
Membrane lipid composition modulates the organization of VDAC1, a mitochondrial gatekeeper
Elodie Lafargue, Jean-Pierre Duneau, Nicolas Buzhinsky, Pamela Ornelas, Alexandre Ortega, Varun Ravishankar, James Sturgis, Ignacio Casuso, Lucie Bergdoll
Communications Biology 8:936 (2025)10.1038/s42003-025-08311-5
High‐Speed Atomic Force Microscopy Reveals the Dynamic Interplay of Membrane Proteins is Lipid‐Modulated
Eunji Shin, Yining Jiang, Batiste Thienpont, James N. Sturgis, Simon Scheuring
Small Science (2025)10.1002/smsc.202500258
Pushing the Boundaries of Resolution in Solid-State Nuclear Magnetic Resonance of Biomolecules with 160 kHz Magic-Angle Spinning
Zhiyu Sun, Claire Ollier, Adrienn Rancz, Batiste Thienpont, Kristof Grohe, Lukas Becker, Armin Purea, Frank Engelke, Sebastian Wegner, James N. Sturgis, Tatyana Polenova, Tanguy Le Marchand, Guido Pintacuda
Journal of the American Chemical Society 147:19433-19437 (2025)10.1021/jacs.5c02466
Protocol for the production and reconstitution of VDAC1 for functional assays
Grace I Dearden, Varun Ravishankar, Ken-Taro Sakata, Anant K Menon, Lucie Bergdoll
STAR Protocols 5 (2024)10.1016/j.xpro.2024.103240
Lipids influence the oligomeric organization of VDAC1
Elodie Lafargue, Jean-Pierre Duneau, Nicolas Buzhinsky, James Sturgis, Ignacio Casuso, Lucie Bergdoll
Biophysical Journal 123 (2024)10.1016/j.bpj.2023.11.1061
VDAC interactions with lipids and their effect on isoform specificity
Jean-Pierre Duneau, Maria Queralt-Martin, Lucie Bergdoll
Biophysical Journal 122:373a (2023)10.1016/j.bpj.2022.11.2056
Nanodiscs as a novel approach to resolve inter-protein energy transfer within the photosynthetic membrane of purple bacteria
Olivia Fiebig, Dihao Wang, Dvir Harris, Hila Toporik, Yi Ji, Chern Chuang, Muath Nairat, Ashley Tong, John Ogren, Stephanie Hart, Jianshu Cao, James N. Sturgis, Yuval Mazor, Gabriela Schlau-Cohen
Biophysical Journal 122:232a (2023)10.1016/j.bpj.2022.11.1368
The single residue K12 governs highly sensitive voltage gating of the voltage-dependent anion channel
Van Ngo, Maria Queralt-Martin, Farha Khan, Lucie Bergdoll, Jeff Abramson, Sergey Bezrukov, Tatiana Rostovtseva, Sergei Noskov, David Hoogerheide
Biophysical Journal 122:92a (2023)10.1016/j.bpj.2022.11.697
Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria
Dihao Wang, Olivia Fiebig, Dvir Harris, Hila Toporik, Yi Ji, Chern Chuang, Muath Nairat, Ashley Tong, John Ogren, Stephanie Hart, Jianshu Cao, James Sturgis, Yuval Mazor, Gabriela Schlau-Cohen
Proceedings of the National Academy of Sciences of the United States of America 120:e2220477120 (2023)10.1073/pnas.2220477120
Membrane-mediated membrane protein interactions drive membrane protein organization
Yining Jiang, Batiste Thienpont, James N. Sturgis, Jeremy Dittman, Simon Scheuring
Biophysical Journal 121:433a (2022)10.1016/j.bpj.2021.11.608
Membrane-mediated protein interactions drive membrane protein organization
Yining Jiang, Batiste Thienpont, Vinay Sapuru, Richard Hite, Jeremy Dittman, James Sturgis, Simon Scheuring
Nature Communications 13:7373 (2022)10.1038/s41467-022-35202-8
Experimental evidence for long-distance electrodynamic intermolecular forces
Mathias Lechelon, Yoann Meriguet, Matteo Gori, Sandra Ruffenach, Ilaria Nardecchia, Elena Floriani, Dominique Coquillat, Frederic Teppe, Sébastien Mailfert, Didier Marguet, Pierre Ferrier, Luca Varani, James Sturgis, Jeremie Torres, Pettini Marco
Science Advances 8:eabl5855 (2022)10.1126/sciadv.abl5855
The Single Residue K12 Governs the Exceptional Voltage Sensitivity of Mitochondrial Voltage-Dependent Anion Channel Gating
Van A. Ngo, María Queralt-Martín, Farha Khan, Lucie Bergdoll, Jeff Abramson, Sergey m. Bezrukov, Tatiana K Rostovtseva, David P Hoogerheide, Sergei Yu Noskov
Journal of the American Chemical Society 144:14564-14577 (2022)10.1021/jacs.2c03316
Comparison of the Energy-Transfer Rates in Structural and Spectral Variants of the B800–850 Complex from Purple Bacteria
Ashley Tong, Olivia Fiebig, Muath Nairat, Dvir Harris, Marcel Giansily, Aurélia Chenu, James N. Sturgis, Gabriela Schlau-Cohen
Journal of Physical Chemistry B 124:1460-1469 (2020)10.1021/acs.jpcb.9b11899
High-speed atomic force microscopy highlights new molecular mechanism of daptomycin action
Francesca Zuttion, Adai Colom, Stefan Matile, Denes Farago, Frédérique Pompeo, Janos Kokavecz, Anne Galinier, James N. Sturgis, Ignacio Casuso
Nature Communications 11 (2020)10.1038/s41467-020-19710-z
The lipid environment of Escherichia coli Aquaporin Z
Victoria Schmidt, Marlon Sidore, Cherine Bechara, Jean-Pierre Duneau, James N. Sturgis
Biochimica et Biophysica Acta:Biomembranes 1861:431-440 (2019)10.1016/j.bbamem.2018.10.017
Out-of-Equilibrium Collective Oscillation as Phonon Condensation in a Model Protein
Ilaria Nardecchia, Jeremie Torres, Mathias Lechelon, Valeria Giliberti, Michele Ortolani, Philippe Nouvel, Matteo Gori, Yoann Meriguet, Irene Donato, Jordane Preto, Luca Varani, James N. Sturgis, Marco Pettini
Physical Review X 8:031061 (2018)10.1103/physrevx.8.031061
Modifying Styrene-maleic Acid Co-polymer for Studying Lipid Nanodiscs by Direct Fluorescent Labeling
Victoria Schmidt, James N. Sturgis
Bio-protocol 8 (2018)10.21769/BioProtoc.2969
Modifying styrene-maleic acid co-polymer for studying lipid nanodiscs
Victoria Schmidt, James N. Sturgis
Biochimica et Biophysica Acta:Biomembranes 1860:777-783 (2018)10.1016/j.bbamem.2017.12.012
Making Monomeric Aquaporin Z by Disrupting the Hydrophobic Tetramer Interface
Victoria Schmidt, James N. Sturgis
ACS Omega 2:3017-3027 (2017)10.1021/acsomega.7b00261
Lipid perturbation by membrane proteins and the lipophobic effect
James N. Sturgis, Jean-Pierre Duneau, Jonathan Khao
Biochimica et Biophysica Acta:Biomembranes 1859:126-134 (2017)10.1016/j.bbamem.2016.10.014
Making dimers of oligomeric membrane proteins using copper-free click chemistry
James N. Sturgis, Wang Dong
F1000Research 5 (2016)10.12688/f1000research.8676.1
Lipid Environment of Aquaporin Z
Schmidt Victoria, Sidore Marlon, Carrière Fredéric, Duneau Jean-Pierre, James N. Sturgis
Biophysical Journal 110:252a-253a (2016)10.1016/j.bpj.2015.11.1389
The E.coli Aquaporin Z interface : a puzzle?
Victoria Schmidt, Pierre Hubert, James N. Sturgis
Eur. Biophys. J. 44:S217 (2015)
Transmembrane Recognition of the Semaphorin Co-Receptors Neuropilin 1 and Plexin A1 : Coarse-Grained Simulations
Samia Aci-Sèche, Paul Sawma, Pierre Hubert, James N. Sturgis, Dominique Bagnard, Laurent Jacob, Monique Genest, Norbert Garnier
PLoS ONE 9:e97779 (2014)10.1371/journal.pone.0097779
Ultrafast excited state processes in Roseobacter denitrificans antennae: comparison of isolated complexes and native membranes.
Marco Ferretti, Katia Duquesne, James N. Sturgis, Rienk Van Grondelle
Physical Chemistry Chemical Physics 16:26059-66 (2014)10.1039/c4cp02986k
Evidence for new homotypic and heterotypic interactions between transmembrane helices of proteins involved in receptor tyrosine kinase and neuropilin signaling.
Paul Sawma, Lise Roth, Cécile Blanchard, Dominique Bagnard, Gérard Crémel, Emmanuelle Bouveret, Jean-Pierre Duneau, James N. Sturgis, Pierre Hubert
Journal of Molecular Biology 426:4099-111 (2014)10.1016/j.jmb.2014.10.007
The architecture of Rhodobacter sphaeroides chromatophores
Simon Scheuring, Reinat Nevo, Lu-Ning Liu, Stéphanie Mangenot, Dana Charuvi, Thomas Boudier, Valerie Prima, Pierre Hubert, James N. Sturgis, Ziv Reich
Biochimica biophysica acta (BBA) - Bioenergetics 1837:1263-1270 (2014)10.1016/j.bbabio.2014.03.011
Destabilizing Aquaporin Z Assembly: Effects on Structure, Function and Dynamics
Victoria Schmidt, Pierre Hubert, Valerie Prima, James N. Sturgis
Biophysical Journal 108:499a-500a (2014)
Molecular mechanisms of Tau binding to microtubules and its role in microtubule dynamics in live cells.
Gilles Breuzard, Pierre Hubert, Roqiya Nouar, Tiphany De Bessa, François Devred, Pascale Barbier, James N. Sturgis, Vincent Peyrot
Journal of Cell Science 126:2810-9 (2013)10.1242/jcs.120832
Lateral organization of biological membranes: role of long-range interactions.
Jean-Pierre Duneau, James N. Sturgis
Eur. Biophys. J. 42:843-50 (2013)10.1007/s00249-013-0933-x
Structural properties of the tubular appendage spinae from marine bacterium Roseobacter sp. strain YSCB.
A Bernadac, L-F Wu, C-L Santini, C Vidaud, James N. Sturgis, N Menguy, P Bergam, C Nicoletti, T Xiao
Scientific Reports 2:950 (2012)10.1038/srep00950
Characterization of the motion of membrane proteins using high-speed atomic force microscopy
Ignacio Casuso, Jonathan Khao, Mohamed Chami, Perrine Paul-Gilloteaux, Mohamed Husain, Jean-Pierre Duneau, Henning Stahlberg, James N. Sturgis, Simon Scheuring
Nature Nanotechnology 525-529 (2012)10.1038/nnano.2012.109
Complete Lateral and Angular Diffusion and Protein-Protein Interaction Description of a Membrane Protein
Ignacio Casuso, Jean-Pierre Duneau, Mohamed Chami, Perrine Paul-Gilloteaux, Mohamed Husain, Jonathan Khao, Henning Stahlberg, James N. Sturgis, Simon Scheuring
Biophysical Journal 102:413a-414a (2012)10.1016/j.bpj.2011.11.2261
Draft genome sequence of the purple photosynthetic bacterium Phaeospirillum molischianum DSM120, a particularly versatile bacterium.
K Duquesne, Valérie Prima, B Ji, Z Rouy, C Médigue, E Talla, James N. Sturgis
Journal of Bacteriology 194:3559-60 (2012)10.1128/JB.00605-12
Shotgun genome sequence of the large purple photosynthetic bacterium Rhodospirillum photometricum DSM122.
K Duquesne, James N. Sturgis
Journal of Bacteriology 194:2380 (2012)10.1128/JB.00168-12
Molecular origins and consequences of High-800 LH2 in Roseobacter denitrificans.
Katia Duquesne, Cecile Blanchard, James N. Sturgis
Biochemistry 50:6723-9 (2011)10.1021/bi200538j
Structure of a protein-detergent complex: the balance between detergent cohesion and binding.
Jonathan Khao, Jaime Arce-Lopera, James N. Sturgis, Jean-Pierre Duneau
Eur. Biophys. J. 40:1143-55 (2011)10.1007/s00249-011-0745-9
Native architecture of the photosynthetic membrane from Rhodobacter veldkampii.
Lu-Ning Liu, James N. Sturgis, Simon Scheuring
Journal of Structural Biology 173:138-45 (2011)10.1016/j.jsb.2010.08.010
Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli.
Astrid Wahl, Laetitia My, Romain R. Dumoulin, James N. Sturgis, Emmanuelle Bouveret
Mol. Microbiol. 80:1260-75 (2011)10.1111/j.1365-2958.2011.07641.x
The effects of protein crowding in bacterial photosynthetic membranes on the flow of quinone redox species between the photochemical reaction center and the ubiquinol-cytochrome c2 oxidoreductase.
Kamil Woronowicz, Daniel Sha, Raoul N Frese, James N. Sturgis, Vikas Nanda, Robert A Niederman
Metallomics 3:765-74 (2011)10.1039/c1mt00034a
High-resolution architecture of the outer membrane of the Gram-negative bacteria Roseobacter denitrificans.
Szymon Jarosławski, Katia Duquesne, James N. Sturgis, Simon Scheuring
Molecular Microbiology 74:1211-1222 (2009)10.1111/j.1365-2958.2009.06926.x
Rows of ATP Synthase Dimers in Native Mitochondrial Inner Membranes
Nikolay Buzhynskyy, Pierre Sens, Valérie Prima, James N. Sturgis, Simon Scheuring
Biophysical Journal 93:2870-2876 (2007)10.1529/biophysj.107.109728
Confined diffusion in tubular structures analyzed by fluorescence correlation spectroscopy on a mirror
E. Etienne, P.-F. Lenne, James N. Sturgis, H. Rigneault
Applied optics 45:4497-4507 (2006)10.1364/AO.45.004497
The photosynthetic apparatus of Rhodopseudomonas palustris: Structures and organization.
Simon Scheuring, Rui Pedro Goncalves, Valérie Prima, James N. Sturgis
Journal of Molecular Biology in press:in press (2006)
The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB
E. Cascales, Roland Lloubès, James N. Sturgis
Molecular Microbiology 42:795-807 (2001)10.1046/j.1365-2958.2001.02673.x
Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli
E. Cascales, Marthe Gavioli, James N. Sturgis, Roland Lloubes
Molecular Microbiology 38:904-915 (2000)10.1046/j.1365-2958.2000.02190.x
1H-13C nuclear magnetic resonance assignment and structural characterization of HIV-1 Tat protein.
Jean-Marie Peloponese, C. Gregoire, S Opi, E Esquieu, James N. Sturgis, E Lebrun, E. Meurs, C Collette, D Olive, M Aubertin, M Witvrow, C Pannecouque, E de Clercq, Cedric Bailly, J. Lebreton, E. Loret
Comptes Rendus de l'Académie des Sciences - Series III - Sciences de la Vie 323:883-94 (2000)10.1016/S0764-4469(00)01228-2
Collaborations
- National
- Olivia du Roure, ESPCI ParisTech
- Julien Heuvingh, ESPCI ParisTech
- Clément Campillo, Université d’Evry
- Guido Pintacuda, CNRS ISA, Lyon
- Marco Pettini, CNRS CPT, Marseille
- Ignacio Casuso, INSERM, Marseille
- International
- David Kovar University of Chicago, USA
- Robert Robinson IMCB, Singapore
- Simon Scheuring, Corning Medical School, USA
- Janice Robertson, Washington University, USA
- Gabriela Schlau-Cohen, MIT, USA
- Alexandra Olaya-Castro, UCL, UK
- Luca Sapienza, Southampton University, UK
Funding














