Our team aims to understand the assembly and function of conserved macrocomplexes present in the envelope of Gram-negative bacteria: the Tol and Ton systems. These protein assemblies, involved in cell adaptation and survival, are also the gateway for bacterial toxins and filamentous phages.
Both Tol and Ton systems harness the energy produced from the proton motive force generated at the inner membrane to transduce it at the outer membrane. These complexes are involved in functions that are essential for bacterial survival.

The Ton system mediates the active uptake across the outer membrane of metal, cyanocobalamin and carbohydrates. The Tol system has a more complex function, as it is involved in outer membrane integrity, in the correct localization of some polar factors and chemoreceptors, in outer-membrane phospholipid homeostasis, and in the constriction of the outer membrane during cell division.
Both Tol and Ton systems can be parasitized to open a gate for bacteriotoxins (called colicins), which result in bacterial cell death. These systems are also the target of filamentous phages during the infection process, such as the M13 phage targeting E. coli, and the CTX phage which is specific of the human pathogen Vibrio cholerae.
To obtain an integrate view of the inner membrane Tol and Ton complexes, we developpe an interdisciplinary study in collaboration with different laboratories, mixing genetic, biochemical and microscopy techniques.
1 – Structural and functional analysis of the Tol and Ton complexes.
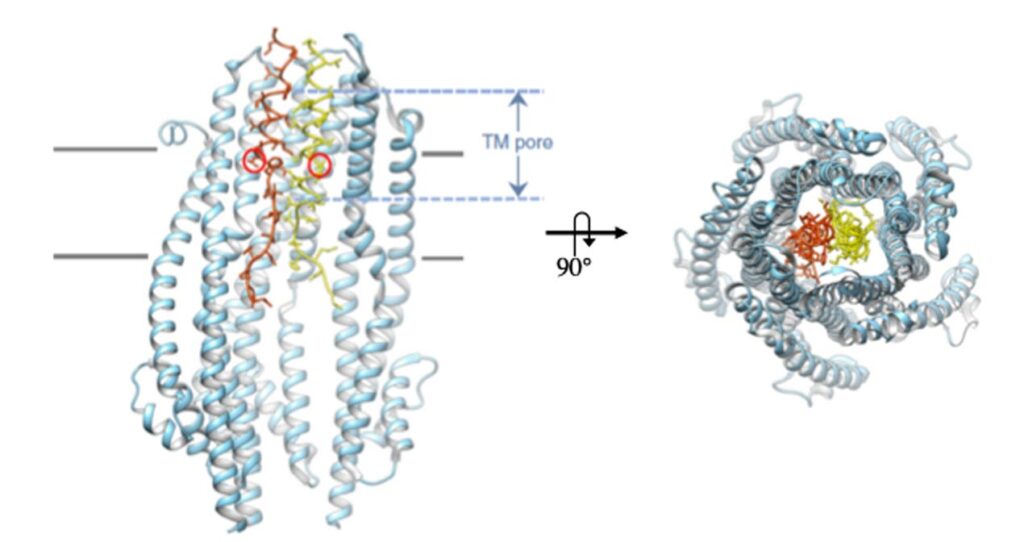
Structural and functional studies on the Tol and Ton complexes isolated from various Gram negative bacteria are investigated in collaboration with international teams. More specifically, we are interested in the interactions occuring between the complex subunits in vivo, the structural study of soluble domain by NMR, the structural and dynamic studies of the complexes by EPR and the high-resolution structure of the membrane complexes.
2 – Dynamic study of the Tol-Pal proteins
We have started to develop the in vivo imaging techniques of the Tol-Pal proteins in different background strains using fluorescence microscopy. In collaboration, we use high resolution fluorescent microscopy and microfluidic device to follow the dynamic of the Tol-Pal proteins. The analyses are performed in relation with cell division, environmental conditions and using specific mutations.

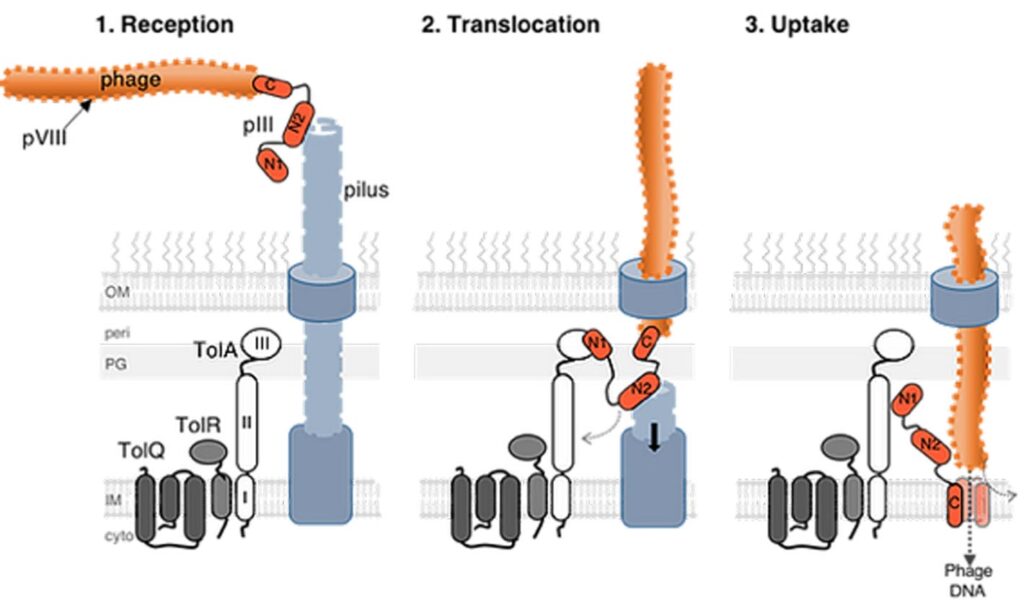
3 – Filamentous phage transport across the envelope and role in host pathogenicity
The Tol system is the entrance gate for a family of bacterial viruses named filamentous phages. For some bacterial species, such as Pseudomonas aeruginosa or Vibrio cholerae, mutualistic association with these phages results in increased pathogenicity and virulence.
Our goal is to identify the determinants of phage-host-specific recognition and to characterize the sequence of molecular events involved during phage transport across the periplasm. Our models are the fd phage infecting E. coli, as well as the CTX and Pf phages, associated with the virulence of their hosts, V. cholerae and P. aeruginosa, respectively.
The expected results will shed new light on the mechanism of phage-dependent pathogenic conversion in bacteria and open new perspectives in the engineering of hybrid phages designed to deliver molecules of interest to pathogens.
Members
Researchers
- Roland Lloubès – Former team leader (DR, CNRS Emeritat)
Technical Staff
- Pauline BACCELLI (ITA, 2019-2021)
- Bastien SERRANNO (Engineer, 2015-2017)
- Hervé CELIA (Engineer CNRS, 2008-2018), now at NIDDK, NIH (USA)
- Marthe GAVIOLI (TCE, retired in 2010)
Ph.D students
- Callypso PELLEGRI (2021-2024) – Analyse fonctionnelle du système Tol-Pal au cours de l’infection par les phages filamenteux.
- Mélissa PETITI (2015-2018) – Assemblage et dynamique in vivo du système Tol-Pal
- Xiang ZHANG (2007-2010) – Organisation fonctionnelle d’un moteur moléculaire Tol et d’une protéine active contre une toxine bactérienne
- Nadir SEDDIKI (codirection Dr Susan Buchanan, NIH, Bethesda) (2007-2010) – Import of macromolecules: Structural studies of the pesticin toxin and of an engineered variant
- Aurélie Barnéoud-Arnoulet (2007-2010)- Mécanismes d’importation des collicines: détournement des fonctions physiologiques des protéines FtsH et FkpA
- Emilie GOEMAERE (2004-2008) – Le complexe membranaire Tol d’E.coli: un modèle pour l’étude de l’organisation fonctionnelle d’un moteur moléculaire bacterien
- Stephanie POMMIER (2002-2005) – Le système Tol-Pal d’E. coli
Post-Doctorants
- Laetitia HOUOT (2010-2012)
- Xiang ZHANG (2011)
Master 2
- Samima RAZIA (2025) – Solution monitoring of the conformational changes of the TolA mechanical arm
- Romane GUARINO (2024) – Caractérisation de la dynamique de l’infection bactérienne par les phages filamenteux à l’échelle de la cellule unique.
- Identification des étapes tardives de la division cellulaire chez les bactéries à Gram négatif : Rôle du système Tol-Pal.
- Callypso PELLEGRI (2021) – Analyse fonctionnelle du système Tol-Pal au cours de l’infection par les phages filamenteux.
- Ambre MOREAU (2020) – Mécanisme moléculaire d’infection par les phages filamenteux: réseau d’interactions protéiques mis en jeu.
- Melvin RENAULT (2019) – Etude de la localisation des moteurs Tol et Ton et de l’interchangeabilité des moteurs chez E. coli.
- Poutoum SAMIRE (2018) – Importance du moteur énergisé TolQRA et de la flexibilité de TolA dans l’import des phages filamenteux.
- Nicolas GOMEZ (2016) – Analyse fonctionnelle du système Tol-Pal en fonction de l’état redox de TolA
- Laura FRADALE (2015) – Analyse transcriptionnelle et fonctionnelle du système Tol-Pal chez Vibrio cholera
- Melissa PETITI (2014) – Etude du système Tol-Pal par mutagénèse dirigée et double hybride bactérien
- Perinne VAN OVERTVELT (2011)
- Xiang ZHANG (2007)
- Aurélie BARNEOUD-ARNOULET (2007)
Master 1
- Khadidja OUENZAR (2024)
- Wajdi JAOUAD 2023
- Lara PELZER (2022)
- Léa MOURIER (2022)
- Rachida SMIMIH (2022)
- Megan CHARLES-ALFRED (2019)
- Emeline LEMARIE (2019)
- Melvin RENAULT (2018)
- Poutoum SAMIRE (2017)
- Raphaël RACHEDI (2017)
- Pauline BACCELLI (2014)
- Marie DESCAMPS (2014)
- Alexandra LIPSKI (2006)
Undergraduates
- Cloé BARTHELEMI, BTS (2023)
- Virgile LEBORGNE, 3ème (2023)
- Andréa BORGOGNO-BERTI, 3ème (2023)
- Ally PONTET (2023)
- Alice OLIVIER, L2 (2022)
- Chiama BOUALI, BTS (2022)
- Elsa PONCET, L3 Lyon (2022)
- Léonie DELEUIL, BUT Toulon (2022)
- Maxime RACCHINI, L2 (2022)
- Maximilien ROUZAUD, L3 (2022)
- Lola JUDOR, L2 (2022)
- Garance CHABANNES (UE Labo in vivo, L2, 2012)
- Chaima BOUDI, BTS (2022)
- Méline SEEL, BTS (2020)
- Jennifer GONCALVES DA SILVA (UE Labo in vivo, L2, 2019)
- Myriam BOUSSOUFA (UE Labo in vivo, L2, 2019)
- Defne AKSOY (UE Labo in vivo, L2, 2019)
- Celia JONAS (UE Labo in vivo, L2, 2019)
- Victoria SGOLUPPI, L3 (2018)
- Faniry RABOANARIVOLA, L3 (2017)
- Jad MOUALLEM, UE Labo in vivo L1 (2017)
- Megan CHARLES-ALFRED, L3 (2017)
- Ahmed NASRAOUI, UE Labo in vivo L1 (2016)
- Clara FERNANDEZ, L2 (2016)
- Laura BASSET, ESIL-Polytech (2015)
- Marie-Sarah ROUVIERE, UE Labo in vivo L1 (2015)
- Axel DEJARDIN, BTS (2014, 2015)
- Lia FRIEIER, ERASMUS L3 (2013)
- Audrey FAVARO, ESIL-Polytech (2012)
- Clement NOEL, ESIL-Polytech (2012)
- Stéphanie ROLLERO, ESIL-Polytech (2010)
- Sarah DOCHE, ESIL-Polytech (2009)
- Nathalie MIARD, INSA Toulouse (2007)
- Patrick ROCKENFELLER, ERASMUS (2005)
Publications
Direct interaction between fd phage pilot protein pIII and the TolQ–TolR proton-dependent motor provides new insights into the import of filamentous phages
Callypso Pellegri, Ambre Moreau, Denis Duché, Laetitia Houot
Journal of Biological Chemistry 299:105048 (2023)10.1016/j.jbc.2023.105048
Timing of TolA and TolQ recruitment at the septum depends on the functionality of the Tol-Pal system
Pauline Baccelli, Raphaël Rachedi, Bastien Serrano, Mélissa Petiti, Christophe Bernard, Laetitia Houot, Denis Duché, Denis Duche #
Journal of Molecular Biology 434:167519 (2022)10.1016/j.jmb.2022.167519
The Tol-Pal system of Escherichia coli plays an unexpected role in the import of the oxyanions chromate and phosphate
Amine Ali Chaouche, Laetitia Houot, Denis Duché, Chantal Iobbi-Nivol, Marie-Thérèse Giudici-Orticoni, Michel Fons, Vincent Méjean
Research in Microbiology 173:103967 (2022)10.1016/j.resmic.2022.103967
Fur -Dam regulatory interplay at an internal promoter of the enteroaggregative Escherichia coli Type VI secretion sci1 gene cluster
Yannick R Brunet, Christophe S Bernard, E. Cascales
Journal of Bacteriology (2020)
Connected partner-switches control the life style of Pseudomonas aeruginosa through RpoS regulation
Sophie Bouillet, Moly Ba, Laetitia Houot, Chantal Iobbi-Nivol, Christophe Bordi
Scientific Reports 9 (2019)10.1038/s41598-019-42653-5
Similarities and Differences between Colicin and Filamentous Phage Uptake by Bacterial Cells
Denis Duché, Laetitia Houot
EcoSal Plus 8 (2019)10.1128/ecosalplus.ESP-0030-2018
Tol Energy-Driven Localization of Pal and Anchoring to the Peptidoglycan Promote Outer-Membrane Constriction
Melissa Petiti, Bastien Serrano, Laura Faure, Roland Lloubes, Tam Mignot, Denis Duché
Journal of Molecular Biology 431:3275-3288 (2019)10.1016/j.jmb.2019.05.039
Decoupling filamentous phage uptake and energy of the TolQRA motor in Escherichia coli .
Poutoum Samire, Bastien Serrano, Denis Duché, Emeline Lemarie, Roland Lloubes, Laetitia Houot
Journal of Bacteriology (2019)10.1128/JB.00428-19
Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: Intracellular delivery, trafficking and mechanisms of cell injury
Martina Bielaszewska, Christian Rüter, Andreas Bauwens, Lilo Greune, Kevin-André Jarosch, Daniel Steil, Wenlan Zhang, Xiaohua He, Roland Lloubès, Angelika Fruth, Kwang Sik K. S. Kim, Alexander Schmidt, Ulrich Dobrindt, Alexander Mellmann, Helge Karch
PLoS Pathogens 13 (2017)10.1371/journal.ppat.1006159
Electrostatic interactions between the CTX phage minor coat protein and the bacterial host receptor TolA drive the pathogenic conversion of Vibrio cholerae
Laetitia Houot, Romain Navarro, Matthieu Nouailler, Denis Duché, Francoise Guerlesquin, Roland Lloubès
Journal of Biological Chemistry 292:13584-13598 (2017)10.1074/jbc.M117.786061
Structural insight into the role of the Ton complex in energy transduction
Hervé H. Celia, Nicholas Noinaj, Stanislav D Zakharov, Enrica Bordignon, Istvan Botos, Mónica Santamaria, Travis J Barnard, William A Cramer, Roland Lloubès, Susan K Buchanan
Nature 538:60-65 (2016)10.1038/nature19757
(1)H, (15)N and (13)C resonance assignments of the C-terminal domain of Vibrio cholerae TolA protein.
Romain Navarro, Olivier Bornet, Laetitia Houot, Roland Lloubes, Francoise Guerlesquin, Matthieu Nouailler
Biomol NMR Assign (2016)10.1007/s12104-016-9690-y
Virulence from vesicles: Novel mechanisms of host cell injury by Escherichia coli O104:H4 outbreak strain
Lisa Kunsmann, Christian Rüter, Andreas Bauwens, Lilo Greune, Malte Glüder, Björn Kemper, Angelika Fruth, Sun Nyunt Wai, Xiaohua He, Roland Lloubes, M. Alexander Schmidt, Ulrich Dobrindt, Alexander Mellmann, Helge Karch, Martina Bielaszewska
Scientific Reports 1-18 (2015)10.1038/srep13252
Fluorescence resonance energy transfer based on interaction of PII and PipX proteins provides a robust and specific biosensor for 2-oxoglutarate, a central metabolite and a signalling molecule.
Hai-Lin Chen, Christophe S Bernard, Pierre Hubert, Laetitia My, Cheng-Cai Zhang
FEBS Journal 281:1241-55 (2014)10.1111/febs.12702
Non classical secretion systems.
Roland Lloubes, Alain Bernadac, Laetitia Houot, Stéphanie Pommier
Res. Microbiol. 164:655-63 (2013)10.1016/j.resmic.2013.03.015
Mannitol and the mannitol-specific enzyme IIB subunit activate Vibrio cholerae biofilm formation.
Patrick Ymele-Leki, Laetitia Houot, Paula I Watnick
Applied and Environmental Microbiology 79:4675-83 (2013)10.1128/AEM.01184-13
Characterization of colicin M and its orthologs targeting bacterial cell wall peptidoglycan biosynthesis.
Hélène Barreteau, Meriem El Ghachi, Aurélie Barnéoud-Arnoulet, Emmanuelle Sacco, Thierry Touzé, Denis Duché, Fabien Gérard, Mark Brooks, Delphine Patin, Ahmed Bouhss, Didier Blanot, Herman Van Tilbeurgh, Michel Arthur, Roland Lloubes, Dominique Mengin-Lecreulx
Microb. Drug Resist. 18:222-9 (2012)10.1089/mdr.2011.0230
A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa.
Laetitia Houot, Adeline Fanni, Sophie De Bentzmann, Christophe Bordi
Microbiology (Reading, Engl.) 158:1964-71 (2012)10.1099/mic.0.057059-0
Energetics of colicin import revealed by genetic cross-complementation between the Tol and Ton systems.
Roland Lloubes, Emilie Goemaere, Xiang Y. Zhang, E. Cascales, Denis Duché
Biochem. Soc. Trans. 40:1480-5 (2012)10.1042/BST20120181
Energetics of colicin import revealed by genetic cross-complementation between the Tol and Ton systems
Roland Lloubès, Emilie Goemaere, Xiang Zhang, E. Cascales, Denis Duché
Biochemical Society Transactions 40:1480-1485 (2012)10.1042/BST20120181
Colicin M, a peptidoglycan lipid-II-degrading enzyme: potential use for antibacterial means?
Thierry Touzé, Hélène Barreteau, Meriem El Ghachi, Ahmed Bouhss, Aurélie Barnéoud-Arnoulet, Delphine Patin, Emmanuelle Sacco, Didier Blanot, Michel Arthur, Denis Duché, Roland Lloubes, Dominique Mengin-Lecreulx
Biochem. Soc. Trans. 40:1522-7 (2012)10.1042/BST20120189
The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 Type VI secretion system, is inserted by YidC.
Abdelrahim Zoued, Roland Lloubes, Laure Journet, E. Cascales, Marie-Stéphanie Aschtgen
MicrobiologyOpen 1:71-82 (2012)10.1002/mbo3.9
Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins.
Christophe S Bernard, Yannick R Brunet, Marthe Gavioli, Roland Lloubes, E. Cascales
Journal of Bacteriology 193:2158-67 (2011)10.1128/JB.00029-11
An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster
Yannick R Brunet, Christophe S Bernard, Marthe Gavioli, Roland Lloubes, E. Cascales
PLoS Genetics 7:e1002205 (2011)10.1371/journal.pgen.1002205
The anaerobe-specific orange protein complex of Desulfovibrio vulgaris hildenborough is encoded by two divergent operons coregulated by ?54 and a cognate transcriptional regulator.
Anouchka Fiévet, Laetitia My, E. Cascales, Mireille Ansaldi, Sofia R Pauleta, Isabel Moura, Zorah Dermoun, Christophe S Bernard, Alain Dolla, Corinne Aubert
Journal of Bacteriology 193:3207-19 (2011)10.1128/JB.00044-11
Mapping the interactions between Escherichia coli TolQ transmembrane segments
Xiang Y-Z Zhang, Emilie L Goemaere, Nadir Seddiki, Hervé H. Celia, Marthe Gavioli, E. Cascales, Roland Lloubes
Journal of Biological Chemistry 286:11756-64 (2011)10.1074/jbc.M110.192773
Toxicity of the Colicin M Catalytic Domain Exported to the Periplasm Is FkpA Independent
Aurélie Barnéoud-Arnoulet, Hélène Barreteau, Thierry Touze, Dominique Mengin-Lecreulx, Roland Lloubes, Denis Duché
Journal of Bacteriology 192:5212-5219 (2010)10.1128/JB.00431-10
Deciphering the Catalytic Domain of Colicin M, a Peptidoglycan Lipid II-degrading Enzyme
Hélène Barreteau, Ahmed Bouhss, Fabien Gérard, Denis Duché, Boubekeur Boussaid, Didier Blanot, Roland Lloubes, Dominique Mengin-Lecreulx, Thierry Touze
Journal of Biological Chemistry 285:12378-12389 (2010)10.1074/jbc.M109.093583
Determinants of the Proton Selectivity of the Colicin A Channel
Stephen Slatin, Daniel Baty, Denis Duché
Biochemistry 49:4786-4793 (2010)10.1021/bi100122g
Channel Domain of Colicin A Modifies the Dimeric Organization of Its Immunity Protein
Xiang Y-Z Zhang, Roland Lloubes, Denis Duché
Journal of Biological Chemistry 285:38053-38061 (2010)10.1074/jbc.M110.144071
Immunity Protein Protects Colicin E2 from OmpT Protease
Denis Duché, Mohamed Issouf, Roland Lloubes
Journal of Biochemistry 145:95-101 (2008)10.1093/jb/mvn149
Colicin biology.
E. Cascales, Susan Buchanan, Denis Duché, Colin Kleanthous, Roland Lloubes, Kathleen Postle, Margaret Riley, Stephen Slatin, Danièle Cavard
Microbiology and Molecular Biology Reviews 71:158-229 (2007)10.1128/MMBR.00036-06
Colicin E2 Is Still in Contact with Its Receptor and Import Machinery When Its Nuclease Domain Enters the Cytoplasm
Denis Duché
Journal of Bacteriology 189:4217-4222 (2007)10.1128/JB.00092-07
Release of Immunity Protein Requires Functional Endonuclease Colicin Import Machinery
Denis Duché, Aurélie Frenkian, Valérie Prima, Roland Lloubes
Journal of Bacteriology 188:8593-8600 (2006)10.1128/JB.00941-06
Combining high-field EPR with site-directed spin labeling reveals unique information on proteins in action
K. Möbius, A. Savitsky, C. Wegener, M. Plato, M. Fuchs, A. Schnegg, A. Dubinskii, Y. Grishin, I. Grigor'Ev, M. Kühn, Denis Duché, H. Zimmermann, H.-J. Steinhoff
Magnetic Resonance in Chemistry 43:S4-S19 (2005)10.1002/mrc.1690
Improved methods for producing outer membrane vesicles in Gram-negative bacteria.
Thomas Henry, Stéphanie Pommier, Laure Journet, Alain Bernadac, Jean-Pierre Gorvel, Roland Lloubès
Research in Microbiology 155:437-46 (2004)10.1016/j.resmic.2004.04.007
Spontaneous Refolding of the Pore-Forming Colicin A Toxin upon Membrane Association As Studied by X-Band and W-Band High-Field Electron Paramagnetic Resonance Spectroscopy †
Anton Savitsky, Martin Kühn, Denis Duché, Klaus Möbius, Heinz-Jürgen Steinhoff
Journal of Physical Chemistry B 108:9541-9548 (2004)10.1021/jp036397l
Gating Movements of Colicin A and Colicin Ia Are Different
S.L. Slatin, Denis Duché, P.K. Kienker, D. Baty
Journal of Membrane Biology 202:73-83 (2004)10.1007/s00232-004-0720-9
Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains.
Emmanuelle Bouveret, Laure Journet, Anne Walburger, E. Cascales, Hélène Bénédetti, Roland Lloubes
Biochimie 84:413-21 (2002)
The pore-forming domain of colicin A fused to a signal peptide: a tool for studying pore-formation and inhibition
Denis Duché
Biochimie 84:455-464 (2002)10.1016/s0300-9084(02)01424-4
Colicin A Immunity Protein Interacts with the Hydrophobic Helical Hairpin of the Colicin A Channel Domain in the Escherichia coli Inner Membrane
Angèle Nardi, Yves Corda, Daniel Baty, Denis Duché
Journal of Bacteriology 183:6721-6725 (2001)10.1128/JB.183.22.6721-6725.2001
The C-terminal half of the colicin A pore-forming domain is active in vivo and in vitro11Edited by I. B. Holland
Angèle Nardi, Stephen Slatin, Daniel Baty, Denis Duché
Journal of Molecular Biology 307:1293-1303 (2001)10.1006/jmbi.2001.4524
Integration of the colicin A pore-forming domain into the cytoplasmic membrane of Escherichia coli 1 1Edited by I. B. Holland
Denis Duché, Yves Corda, Vincent Géli, Daniel Baty
Journal of Molecular Biology 285:1965-1975 (1999)10.1006/jmbi.1998.2423
Intracellular Immunization of Prokaryotic Cells against a Bacteriotoxin
Patrick Chames, Jacques Fieschi, Daniel Baty, Denis Duché
Journal of Bacteriology 180:514-518 (1998)10.1128/JB.180.3.514-518.1998
The channel domain of colicin A is inhibited by its immunity protein through direct interaction in the Escherichia coli inner membrane.
D Espesset, Denis Duché, D. Baty, V. Geli
EMBO Journal 15:2356-64 (1996)
Quantification of group A colicin import sites.
Denis Duché, L. Letellier, V Géli, H Bénédetti, D. Baty
Journal of Bacteriology 177:4935-4939 (1995)10.1128/jb.177.17.4935-4939.1995
Uncoupled steps of the colicin A pore formation demonstrated by disulfide bond engineering.
Denis Duché, M Parker, J González-Mañas, F. Pattus, D. Baty
Journal of Biological Chemistry 269:6332-9 (1994)
Unfolding of colicin A during its translocation through the Escherichia coli envelope as demonstrated by disulfide bond engineering.
Denis Duché, D. Baty, M. Chartier, L. Letellier
Journal of Biological Chemistry 269:24820-5 (1994)10.1016/S0021-9258(17)31465-5
The colicin A pore-forming domain fused to mitochondrial intermembrane space sorting signals can be functionally inserted into the Escherichia coli plasma membrane by a mechanism that bypasses the Tol proteins
David Espesset, Yves Corda, Kyle Cunningham, Hélène Bénédetti, Roland Lloubes, Claude Lazdunski, Vincent Géli
Molecular Microbiology 13:1121-1131 (1994)10.1111/j.1365-2958.1994.tb00503.x
A single amino acid substitution can restore the solubility of aggregated colicin A mutants in Escherichia coli
Jacques Izard, Michael Parker, Martlne Chartier, Denis Duché, Daniel Baty
Protein Engineering, Design and Selection 7:1495-1500 (1994)10.1093/protein/7.12.1495
Fluorescence Energy Transfer Distance Measurements
Jeremy Lakey, Denis Duché, Juan-Manuel Gonzáles-Mañas, Daniel Baty, Franc Pattus
Journal of Molecular Biology 230:1055-1067 (1993)10.1006/jmbi.1993.1218
Collaborations
- National
- T. Mignot, LCB UMR7283, Marseille
- International
- S. Buchanan, NIDDK, NIH, Bethesda, MD, USA
Grants
- IM2B Interdisciplinarité (2022-2024)
- ANR MeMox – Projet ANR- 18-CE11-0027 (2018-2021)
- ANR BacMolMot – Projet ANR-14-CE09-0023 (2014-2017)
- FRM Postdoctoral fellowship (2010-2012)
- ANR Sodatol – Projet ANR- 07-BLAN-067
Funding