
Our team aims to unravel the modus operandi of viruses to recognize their hosts, inactivate cellular defenses and replicate their genome, by combining structural biology, biochemistry and cellular biology.
We are working on 1) bacteriophages infecting lactic acid bacteria (Oenococcusoeni, Lactococcus lactis, Streptococcus thermophilus) and mycobacteria (Mycobacterium smegmatis, Mycobacterium tuberculosis, Mycobacterium abscessus), and 2) human and animal RNA viruses (SARS-CoV-2 and Hepatitis E virus).
Deploying multidisciplinary approach, we aim to define the mechanisms of viral genome replication.
1-Bacteriophages
Bacteriophages (phages) are the most abundant and diverse biological entities on Earth. Yet, several aspects of their biology remain remarkably elusive. Our main research interest is to unravel the molecular mechanisms used by phages in the early steps of infection to recognize and bind to their specific hosts, and to inactivate the bacterial immune system CRISPR-Cas9. This will provide valuable fundamental knowledge to better understand phage-bacteria interactions and to pave the way for the rational development of phage-based applications for biocontrol, diagnostic and therapeutic purposes.
Host binding machineries
The first step in phage infection is recognition and binding to a specific host cell. Phages’ host-binding machineries are multiprotein nanodevices evolved to adapt to cell surface receptors. We and others have shown that siphophages’ host-binding machineries that bind to lactic acid bacteria cell wall polysaccharides are LEGO®-like assemblies highly diverse in their composition, architecture and function. Moreover, we have identified unique features in the host-binding machineries of phages infecting mycobacteria, which are likely adapted to the peculiar composition of their host cell wall.
What is the structural and functional diversity of host-binding devices?
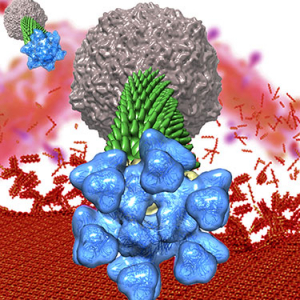
Anti-CRISPR-Cas9 proteins
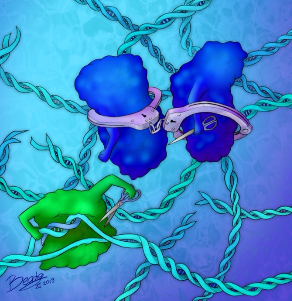
The endless battle for survival between phages and their hosts drives an evolutionary arms race with co-adaptation of their attack and defense mechanisms. In particular, phages have evolved highly diverse anti-CRISPR-Cas9 proteins (Acrs) to inactivate the bacterial CRISPR-Cas9 adaptative immunity and proliferate. We have described the molecular mechanism of AcrIIA6, an Acr identified in S. thermophilus virulent phages, which was the first example of a Cas9 allosteric inhibitor preventing cleavage of target DNA (the last step of CRISPR-Cas9 immunity). Interestingly, the first example of an Acr inhibiting the integration of viral DNA into the bacterial chromosome (the first step of CRISPR-Cas9 immunity) is a short version of AcrIIA6. Yet, the molecular mode of action of this truncated Acr remains unknown.
What is the structural and functional diversity of anti-CRISPR-Cas9 proteins.
Allosteric inhibitor that affects Cas9 structure and dynamics associated to DNA binding.
2-Human and animal RNA viruses
RNA viruses are responsible for many (re-)emerging infectious diseases (Ebola, Zika, Dengue, Coronaviruses…). Unfortunately, most of the time, we have few weapons to fight them, due to a certain lack of detailed knowledge of their propagation cycle. Combining interdisciplinary approaches, we aim to dissect fascinating RNA virus replication machineries. These data will lay the groundwork for the discovery of new antiviral molecules. Indeed, viral replicases have already proved their worth with, as example, the development of therapies against HIV and HCV.
SARS-CoV-2 replicative machinery dissection
Interestingly, among (+) RNA viruses, coronaviruses stand out as having the largest (~30-kb) single-stranded RNA genome known to date associated paradoxically with low mutations rates. CoVs encode a huge replication/transcription machinery consisting, at least, of 16 viral nonstructural proteins (nsps). Thus, a central question is by which mechanisms, these viruses maintain their astonishingly large RNA genome. Based on our past achievement, we aim to characterize the SARS-CoV-2 proofreading machinery.
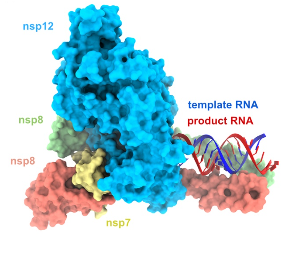
Hepatitis E Virus genome replication mechanisms
Hepatitis E virus (HEV) is a major global health problem and has infected one-third of the world’s population. At least 20 million new infections occur annually resulting in ~70,000 deaths globally. It is the most common cause of acute viral hepatitis but chronic HEV infection, HEV-related acute hepatic failure, and multiple extrahepatic manifestations are now frequently reported. Currently, there is no specific treatment for this virus. While this positive-sense single-stranded RNA genome virus was discovered over 3 decades ago, it remains an enigmatic virus regarding viral life cycle and replication machinery.
Members
Publications
Characterization of strictly lytic phages infecting Oenococcus oeni from Merlot wines and proposal of a new genus
Yasma Barchi, Florencia Oviedo-Hernandez, Amel Chaïb, Cécile Philippe, Olivier Claisse, Christian Cambillau, Adeline Goulet, Claire Le Marrec
Microbiology Spectrum 13 (2025)10.1128/spectrum.02588-24
Structural model of a bacterial focal adhesion complex
Christian Cambillau, Tâm Mignot
Communications Biology 8:119 (2025)10.1038/s42003-025-07550-w
Predicted Structures of Ceduovirus Adhesion Devices Highlight Unique Architectures Reminiscent of Bacterial Secretion System VI
Adeline Goulet, Jennifer Mahony, Douwe van Sinderen, Christian Cambillau
Viruses 17:1261 (2025)10.3390/v17091261
Structure of the T9SS PorKN ring complex reveals conformational plasticity based on the repurposed FGE fold
Xiangan Liu, John D Perpich, Liqiang Song, Christian Cambillau, Thierry Doan, Lei Zheng, Richard J Lamont, Eric Cascales, Bo Hu
mBio 16:e01799-25 (2025)10.1128/mbio.01799-25
Isolation and structure elucidation of cell surface polysaccharides from Oenococcus oeni
Emmanuel Maes, Irina Sadovskaya, Nao Yamakawa, Adeline Goulet, Claire Le Marrec, Marie-Pierre Chapot-Chartier
Carbohydrate Research 552:109456 (2025)10.1016/j.carres.2025.109456
The great phage escape: Activating and escaping lactococcal antiphage systems
Cas Mosterd, Andriana Grafakou, Guillermo Ortiz Charneco, Paul de Waal, Irma van Rijswijck, Noël van Peij, Christine Péchoux, Saulius Kulakauskas, Christian Cambillau, Jennifer Mahony, Douwe van Sinderen
Proceedings of the National Academy of Sciences of the United States of America 122:e2426508122 (2025)10.1073/pnas.2426508122
Lactococcal phage–host profiling through binding studies between cell wall polysaccharide types and Skunavirus receptor-binding proteins
Kelsey White, Giovanni Eraclio, Brian Mcdonnell, Gabriele Andrea Lugli, Tadhg Crowley, Marco Ventura, Federica Volonté, Christian Cambillau, Fabio Dal Bello, Jennifer Mahony, Douwe van Sinderen
Microbial Genomics 11:001395 (2025)10.1099/mgen.0.001395
Looking back at the timely launch of Nature Structural Biology in 1994
Christian Cambillau
Nature Structural and Molecular Biology 31:1812-1812 (2024)10.1038/s41594-024-01436-x
Discovery of antiphage systems in the lactococcal plasmidome
Andriana Grafakou, Cas Mosterd, Matthias H Beck, Philip Kelleher, Brian Mcdonnell, Paul P de Waal, Irma M van Rijswijck, Noël N van Peij, Christian Cambillau, Jennifer Mahony, Douwe van Sinderen
Nucleic Acids Research 52:9760-9776 (2024)10.1093/nar/gkae671
Molecular mechanisms underlying the structural diversity of rhamnose-rich cell wall polysaccharides in lactococci
Hugo Guérin, Pascal Courtin, Alain Guillot, Christine Péchoux, Jennifer Mahony, Douwe van Sinderen, Saulius Kulakauskas, Christian Cambillau, Thierry Touzé, Marie-Pierre Chapot-Chartier
Journal of Biological Chemistry 300:105578 (2024)10.1016/j.jbc.2023.105578
Deciphering the adsorption machinery of Deep-Blue and Vp4, two myophages targeting members of the Bacillus cereus group
Manon Nuytten, Audrey Leprince, Adeline Goulet, Jacques Mahillon
Journal of Virology 98 (2024)10.1128/jvi.00745-24
The tal gene of lactococcal bacteriophage TP901-1 is involved in DNA release following host adsorption
Sofía Ruiz-Cruz, Andrea Erazo Garzon, Christian Cambillau, Guillermo Ortiz Charneco, Gabriele Andrea Lugli, Marco Ventura, Jennifer Mahony, Douwe van Sinderen
Applied and Environmental Microbiology 90:00694-24 (2024)10.1128/aem.00694-24
Combining DEVS simulation and ontological modeling for hierarchical analysis of the SARS-CoV-2 replication
Ali Ayadi, Claudia Frydman, Wissame Laddada, Isabelle Imbert, Cecilia Zanni-Merk, Lina Soualmia
SIMULATION: Transactions of The Society for Modeling and Simulation International (2023)10.1177/00375497231176085
Editorial: The “proteine 2022” conference
Francesco Balestri, Adeline Goulet, Gabriella Tedeschi
Frontiers in Molecular Biosciences 10 (2023)10.3389/fmolb.2023.1157442
Exploring Host-Binding Machineries of Mycobacteriophages with AlphaFold2
Christian Cambillau, Adeline Goulet
Journal of Virology 97:01793-22 (2023)10.1128/jvi.01793-22
Activity and Crystal Structure of the Adherent-Invasive Escherichia coli Tle3/Tli3 T6SS Effector/Immunity Complex Determined Using an AlphaFold2 Predicted Model
Thi Thu Hang Le, Christine Kellenberger, Marie Boyer, Pierre Santucci, Nicolas Flaugnatti, Eric Cascales, Alain Roussel, Stéphane Canaan, Laure Journet, Christian Cambillau
International Journal of Molecular Sciences 24:1740 (2023)10.3390/ijms24021740
Partial Atomic Model of the Tailed Lactococcal Phage TP901-1 as Predicted by AlphaFold2: Revelations and Limitations
Jennifer Mahony, Adeline Goulet, Douwe van Sinderen, Christian Cambillau
Viruses 15 (2023)10.3390/v15122440
Dairy phages escape CRISPR defence of Streptococcus thermophilus via the anti-CRISPR AcrIIA3
Adeline Pastuszka, Geneviève M Rousseau, Vincent Somerville, Sebastien Levesque, Jean-Philippe Fiset, Adeline Goulet, Yannick Doyon, Sylvain Moineau
International Journal of Food Microbiology (2023)
A structural discovery journey of streptococcal phages adhesion devices by AlphaFold2
Adeline Goulet, Raphaela Joos, Katherine Lavelle, Douwe van Sinderen, Jennifer Mahony, Christian Cambillau
Frontiers in Molecular Biosciences 9 (2022)10.3389/fmolb.2022.960325
Exploring Structural Diversity among Adhesion Devices Encoded by Lactococcal P335 Phages with AlphaFold2
Adeline Goulet, Jennifer Mahony, Christian Cambillau, Douwe van Sinderen
Microorganisms 10:2278 (2022)10.3390/microorganisms10112278
Present Impact of AlphaFold2 Revolution on Structural Biology, and an Illustration With the Structure Prediction of the Bacteriophage J-1 Host Adhesion Device
Adeline Goulet, Christian Cambillau
Frontiers in Molecular Biosciences 9:907452 (2022)10.3389/fmolb.2022.907452
Structure Prediction and Analysis of Hepatitis E Virus Non-Structural Proteins from the Replication and Transcription Machinery by AlphaFold2
Adeline Goulet, Christian Cambillau, Alain Roussel, Isabelle Imbert
Viruses 14 (2022)10.3390/v14071537
Replication of the coronavirus genome: A paradox among positive-strand RNA viruses
Emeline Grellet, India l'Hôte, Adeline Goulet, Isabelle Imbert
Journal of Biological Chemistry 298 (2022)10.1016/j.jbc.2022.101923
SARS-CoV-2 detection using a nanobody-functionalized voltammetric device
Quentin Pagneux, Alain Roussel, Hiba Saada, Christian Cambillau, Béatrice Amigues, Vincent Delauzun, Ilka Engelmann, Enagnon Kazali Alidjinou, Judith Ogiez, Anne Sophie Rolland, Julien Poissy, Alain Duhamel, Rabah Boukherroub, David Devos, Sabine Szunerits
Communications Medicine 2:56 (2022)10.1038/s43856-022-00113-8
A truncated anti-CRISPR protein prevents spacer acquisition but not interference
Cécile Philippe, Carlee Morency, Pier-Luc Plante, Edwige Zufferey, Rodrigo Achigar, Denise M Tremblay, Geneviève M Rousseau, Adeline Goulet, Sylvain Moineau
Nature Communications 13 (2022)10.1038/s41467-022-30310-x
Structure and Topology Prediction of Phage Adhesion Devices Using AlphaFold2: The Case of Two Oenococcus oeni Phages
Adeline Goulet, Christian Cambillau
Microorganisms 9:2151 (2021)10.3390/microorganisms9102151
Anchoring the T6SS to the cell wall: Crystal structure of the peptidoglycan binding domain of the TagL accessory protein
van Son Nguyen, Silvia Spinelli, E. Cascales, Alain Roussel, Christian Cambillau, Philippe Leone
PLoS ONE 16:e0254232 (2021)10.1371/journal.pone.0254232
Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9
Daniel Agudelo, Sophie Carter, Minja Velimirovic, Alexis Duringer, Jean-François Rivest, Sébastien Levesque, Jérémy Loehr, Mathilde Mouchiroud, Denis Cyr, Paula Waters, Mathieu Laplante, Sylvain Moineau, Adeline Goulet, Yannick Doyon
Genome Research 30:107-117 (2020)10.1101/gr.255414.119
Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors
Adeline Goulet, Silvia Spinelli, Jennifer Mahony, Christian Cambillau
Viruses 12 (2020)10.3390/v12050512
Diversity of molecular mechanisms used by anti-CRISPR proteins: the tip of an iceberg?
Pierre Hardouin, Adeline Goulet
Biochemical Society Transactions 48:507-516 (2020)10.1042/BST20190638
Revisiting the host adhesion determinants of Streptococcus thermophilus siphophages
Katherine Lavelle, Adeline Goulet, Brian Mcdonnell, Silvia Spinelli, Douwe Van Sinderen, Jennifer Mahony, Christian Cambillau
Microbial Biotechnology 13:1765-1779 (2020)10.1111/1751-7915.13593
Wine phenolic compounds differently affect the host-killing activity of two lytic bacteriophages infecting the lactic acid bacterium oenococcus oeni
Cécile Philippe, Amel Chaib, Fety Jaomanjaka, Stéphanie Cluzet, Aurélie Lagarde, Patricia Ballestra, Alain Decendit, Melina Petrel, Olivier Claisse, Adeline Goulet, Christian C. Cambillau, Claire Le Marrec
Viruses 12:1-19 (2020)10.3390/v12111316
Structural Insights into Lactococcal Siphophage p2 Baseplate Activation Mechanism
Silvia Spinelli, Denise Tremblay, Sylvain Moineau, Christian Cambillau, Adeline Goulet
Viruses 12 (2020)10.3390/v12080878
Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase
Claire Debarnot, Yoan Monneau, Veronique Roig-Zamboni, Vincent Delauzun, Christine Le Narvor, Emeline Richard, Jérôme Hénault, Adeline Goulet, Firas Fadel, Romain R Vivès, Bernard Priem, David Bonnaffé, Hugues Lortat-Jacob, Yves Bourne
Proceedings of the National Academy of Sciences of the United States of America 116:6760-6765 (2019)10.1073/pnas.1818333116
Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6
Olivier Fuchsbauer, Paolo Swuec, Claire Zimberger, Beatrice Amigues, Sébastien Levesque, Daniel Agudelo, Alexis Duringer, Antonio Chaves-Sanjuan, Silvia Spinelli, Geneviève Rousseau, Minja Velimirovic, Martino Bolognesi, Alain Roussel, Christian Cambillau, Sylvain Moineau, Yannick Doyon, Adeline Goulet
Molecular Cell 76:922-937.e7 (2019)10.1016/j.molcel.2019.09.012
Structure–Function Analysis of the C-Terminal Domain of the Type VI Secretion TssB Tail Sheath Subunit
Badreddine Douzi, Laureen Logger, Silvia Spinelli, Stephanie Blangy, Christian Cambillau, E. Cascales
Journal of Molecular Biology 430:297-309 (2018)10.1016/j.jmb.2017.11.015
Towards a complete structural deciphering of Type VI secretion system
van Son Nguyen, Badreddine Douzi, Eric Durand, Alain Roussel, E. Cascales, Christian Cambillau
Current Opinion in Structural Biology 49:77-84 (2018)10.1016/j.sbi.2018.01.007
The gp27-like Hub of VgrG Serves as Adaptor to Promote Hcp Tube Assembly
Melvin G Renault, Jordi Zamarreño Beas, Badreddine Douzi, Maïalène Chabalier, Abdelrahim Zoued, Yannick R Brunet, Christian Cambillau, Laure Journet, E. Cascales
Journal of Molecular Biology 430:3143-3156 (2018)10.1016/j.jmb.2018.07.018
Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics
Badreddine Douzi, Nhung Trinh, Sandra Michel-Souzy, Aline Desmyter, Genevieve Ball, Pascale Barbier, Artemis Kosta, Eric Durand, Katrina Forest, Christian Cambillau, Alain Roussel, Romé Voulhoux
mBio 8 (2017)10.1128/mBio.01185-17
Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex
van Son Nguyen, Laureen Logger, Silvia Spinelli, Pierre Legrand, Thi Thanh Huyen Pham, Thi Trang Nhung Trinh, Yassine Cherrak, Abdelrahim Zoued, Aline Desmyter, Eric Durand, Alain Roussel, Christine Kellenberger, E. Cascales, Christian Cambillau
Nature Microbiology 2:17103 (2017)10.1038/nmicrobiol.2017.103
Characterization of the Porphyromonas gingivalis Type IX Secretion Trans-envelope PorKLMNP Core Complex
Maxence S Vincent, Mickaël J Canestrari, Philippe Leone, Julien Stathopulos, Bérengère Ize, Abdelrahim Zoued, Christian Cambillau, Christine Kellenberger, Alain Roussel, E. Cascales
Journal of Biological Chemistry 292:3252-3261 (2017)10.1074/jbc.M116.765081
TssA: The cap protein of the Type VI secretion system tail
Abdelrahim Zoued, Eric Durand, Yoann G Santin, Laure Journet, Alain Roussel, Christian Cambillau, E. Cascales
BioEssays 39 (2017)10.1002/bies.201600262
Structure and specificity of the Type VI secretion system ClpV-TssC interaction in enteroaggregative Escherichia coli
Badreddine Douzi, Yannick R Brunet, Silvia Spinelli, Valentine Lensi, Stephanie Blangy, Anant Kumar, Laure Journet, Pierre Legrand, E. Cascales, Christian Cambillau
Scientific Reports 6 (2016)10.1038/srep34405
A phospholipase A 1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery
Nicolas Flaugnatti, Thi Thu Hang Le, Stéphane Canaan, Marie-Stéphanie Aschtgen, Vân Nguyên, Stephanie Blangy, Christine Kellenberger, Alain Roussel, Christian Cambillau, E. Cascales, Laure Journet
Molecular Microbiology 99:1099-1118 (2016)10.1111/mmi.13292
Priming and polymerization of a bacterial contractile tail structure. 1" 2" 17
Abdelrahim Zoued, Eric Durand, Yannick R Brunet, Silvia Spinelli, Badreddine Douzi, Mathilde Guzzo, Nicolas Flaugnatti, Pierre Legrand, Laure Journet, Rémi Fronzes, Tam Mignot, Christian Cambillau, E. Cascales
Nature 531:59-63 (2016)10.1038/nature17182
Structure–Function Analysis of the TssL Cytoplasmic Domain Reveals a New Interaction between the Type VI Secretion Baseplate and Membrane Complexes
Abdelrahim Zoued, Chloé J Cassaro, Eric Durand, Badreddine Douzi, Alexandre España, Christian Cambillau, Laure Journet, E. Cascales
Journal of Molecular Biology 428:4413-4423 (2016)10.1016/j.jmb.2016.08.030
Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody
van Son Nguyen, Laureen Logger, Silvia Spinelli, Aline Desmyter, Thi Thu Hang Le, Christine Kellenberger, Badreddine Douzi, Eric Durand, Alain Roussel, E. Cascales, Christian Cambillau
PLoS ONE 10:1-14 (2015)
Production, crystallization and X-ray diffraction analysis of a complex between a fragment of the TssM T6SS protein and a camelid nanobody
van Son Nguyen, Silvia Spinelli, Aline Desmyter, Thi Thu Hang Le, Christine Kellenberger, E. Cascales, Christian Cambillau, Alain Roussel
Acta crystallographica Section F : Structural biology communications [2014-...] 71:266-271 (2015)10.1107/S2053230X15000709
Crystallization and preliminary X-ray analysis of the C-terminal fragment of PorM, a subunit of the Porphyromonas gingivalis type IX secretion system
Julien Stathopulos, Christian Cambillau, E. Cascales, Alain Roussel, Philippe Leone
Acta crystallographica Section F : Structural biology communications [2014-...] 71:71-74 (2015)10.1107/S2053230X1402559X
Crystal Structure and Self-Interaction of the Type VI Secretion Tail-Tube Protein from Enteroaggregative Escherichia coli
Badreddine Douzi, Silvia Spinelli, Stéphanie Blangy, Alain Roussel, Eric Durand, Yannick Brunet, E. Cascales, Christian Cambillau
PLoS ONE 9:e86918 (2014)10.1371/journal.pone.0086918
VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors.
Eric Durand, Christian Cambillau, E. Cascales, Laure Journet
Trends Microbiol. 22:498-507 (2014)10.1016/j.tim.2014.06.004
VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors
Eric Durand, Christian Cambillau, E. Cascales, Laure Journet
Trends in Microbiology 22:498-507 (2014)10.1016/j.tim.2014.06.004
Architecture and assembly of the Type VI secretion system
Abdelrahim Zoued, Yannick Brunet, Eric Durand, Marie-Stéphanie Aschtgen, Laureen Logger, Badreddine Douzi, Laure Journet, Christian Cambillau, E. Cascales
Biochimica et Biophysica Acta - Molecular Cell Research 1843:1664-1673 (2014)10.1016/j.bbamcr.2014.03.018
Architecture and assembly of the Type VI secretion system.
Abdelrahim Zoued, Yannick R Brunet, Eric Durand, Marie-Stéphanie Aschtgen, Laureen Logger, Badreddine Douzi, Laure Journet, Christian Cambillau, E. Cascales
Biochim. Biophys. Acta 1843:1664-73 (2014)10.1016/j.bbamcr.2014.03.018
Biochemical and structural characterization of non-glycosylated Yarrowia lipolytica LIP2 lipase
Ahmed Aloulou, Anaïs Bénarouche, Delphine Puccinelli, Silvia Spinelli, Jean-François Cavalier, Christian Cambillau, Frédéric Carrière
European Journal of Lipid Science and Technology 115:429-441 (2013)10.1002/ejlt.201200440
Dissection of the TssB-TssC Interface during Type VI Secretion Sheath Complex Formation
Xiang Y. Zhang, Yannick R Brunet, Laureen Logger, Badreddine Douzi, Christian Cambillau, Laure Journet, E. Cascales
PLoS ONE 8:e81074 (2013)10.1371/journal.pone.0081074
TssK Is a Trimeric Cytoplasmic Protein Interacting with Components of Both Phage-like and Membrane Anchoring Complexes of the Type VI Secretion System
Abdelrahim Zoued, Eric Durand, Cecilia C. Bebeacua, Yannick R Brunet, Badreddine Douzi, Christian Cambillau, E. Cascales, Laure Journet
Journal of Biological Chemistry 288:27031-27041 (2013)10.1074/jbc.M113.499772
Structural biology of type VI secretion systems
E. Cascales, Christian Cambillau
Philosophical Transactions of the Royal Society B: Biological Sciences 367:1102-1111 (2012)10.1098/rstb.2011.0209
Structural biology of type VI secretion systems.
E. Cascales, Christian Cambillau
Philos. Trans. R. Soc. Lond., B, Biol. Sci. 367:1102-11 (2012)10.1098/rstb.2011.0209
Crystal structure of the VgrG1 actin cross-linking domain of the Vibrio cholerae type VI secretion system.
Eric Durand, Estelle Derrez, Gilles Audoly, Silvia Spinelli, Miguel Ortiz-Lombardia, Didier Raoult, E. Cascales, Christian Cambillau
Journal of Biological Chemistry 287:38190-9 (2012)10.1074/jbc.M112.390153
Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems.
Eric Durand, Abdelrahim Zoued, Silvia Spinelli, Paul J H Watson, Marie-Stéphanie Aschtgen, Laure Journet, Christian Cambillau, E. Cascales
Journal of Biological Chemistry 287:14157-68 (2012)10.1074/jbc.M111.338731
Crystal Structure of the VgrG1 Actin Cross-linking Domain of the Vibrio cholerae Type VI Secretion System
Eric Durand, Estelle Derrez, Gilles Audoly, Silvia Spinelli, Miguel Ortiz-Lombardia, Didier Raoult, E. Cascales, Christian Cambillau
Journal of Biological Chemistry 287:38190-38199 (2012)10.1074/jbc.M112.390153
Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates.
Badreddine Douzi, Geneviève Ball, Christian Cambillau, Mariella Tegoni, Romé Voulhoux
Journal of Biological Chemistry 286:40792-801 (2011)10.1074/jbc.M111.294843
Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Escherichia coli Pathovar
Catarina Felisberto-Rodrigues, Eric Durand, Marie-Stéphanie Aschtgen, Stéphanie Blangy, Miguel Ortiz-Lombardia, Badreddine Douzi, Christian Cambillau, E. Cascales
PLoS Pathogens 7:e1002386 (2011)10.1371/journal.ppat.1002386
Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar
Catarina Felisberto-Rodrigues, Eric Durand, Marie-Stéphanie Aschtgen, Stephanie Blangy, Miguel Ortiz-Lombardia, Badreddine Douzi, Christian Cambillau, E. Cascales
PLoS Pathogens 7:e1002386 (2011)10.1371/journal.ppat.1002386
Collaborations
- National
- Jacques Izopet and Sébastien Lhomme, INFINITy Toulouse https://www.infinity.inserm.fr/equipes-de-recherche-2/equipe-6-j-izopet-b-lagane/
- Béatrice Py, LCB Marseille
- https://lcb.cnrs.fr/team/py/
- Sandrine Ollagnier de Choudens, LCBM Grenoble
- https://www.cbm-lab.fr/Pages/BioCat/Sandrine-Ollagnier.aspx
- Vincent Croquette and Jean-François Allemand, LP ENS Paris https://www.lpens.ens.psl.eu/recherche/biophys/equipe-16/
- Claudia Frydman, LIS Marseille
- https://www.lis-lab.fr/mofed/
- Claire Le Hénaff-Le Marrec, ISVV Bordeaux
- https://oenoresearch.u-bordeaux.fr/fr/
- Marie-Pierre Chapot-Chartier, Institut Micalis
- https://www.micalis.fr/poles-et-equipes/pole-adaptation-bacterienne-et-pathogene/dynamique-de-la-paroi-bacterienne-marie-pierre-chapot-chartier
- Stéphane Canaan, LISM Marseille
- https://lism.imm.cnrs.fr/team/lipolysis-bacterial-pathogenicity/
- International
- Sylvain Moineau, Université Laval Québec (Canada)
- https://www.moineau.bcm.ulaval.ca/index.php?id=2
- Jennifer Mahony, University Cork College (Irlande)
- https://www.ucc.ie/en/apc/people/jennifermahony/
- Douwe van Sinderen, University Cork College (Irlande)
- https://www.ucc.ie/en/apc/people/douwevansinderen/
Funding













